1CKT
 
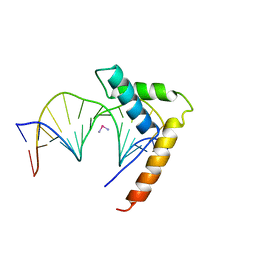 | | CRYSTAL STRUCTURE OF HMG1 DOMAIN A BOUND TO A CISPLATIN-MODIFIED DNA DUPLEX | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*(5IU)P*CP*TP*CP*TP*GP*GP*AP*CP*CP*TP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*GP*TP*CP*CP*AP*GP*AP*GP*AP*GP*G)-3'), ... | | Authors: | Ohndorf, U.-M, Rould, M.A, Pabo, C.O, Lippard, S.J. | | Deposit date: | 1999-04-23 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins.
Nature, 399, 1999
|
|
1D6K
 
 | | NMR SOLUTION STRUCTURE OF THE 5S RRNA E-LOOP/L25 COMPLEX | | Descriptor: | 5S RRNA E-LOOP (5SE), RIBOSOMAL PROTEIN L25 | | Authors: | Stoldt, M, Wohnert, J, Ohlenschlager, O, Gorlach, M, Brown, L.R. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 5S rRNA E-domain-protein L25 complex shows preformed and induced recognition.
EMBO J., 18, 1999
|
|
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2022-02-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3O7L
 
 | |
8GUS
 
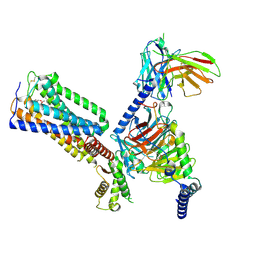 | | Cryo-EM structure of HU-CB2-G protein complex | | Descriptor: | Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
5OJP
 
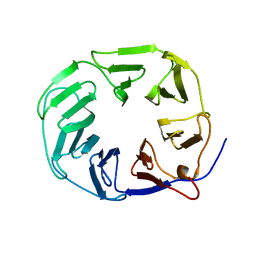 | | YCF48 bound to D1 peptide | | Descriptor: | Ycf48-like protein | | Authors: | Michoux, F, Nixon, P.J, Murray, J.W. | | Deposit date: | 2017-07-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8GUQ
 
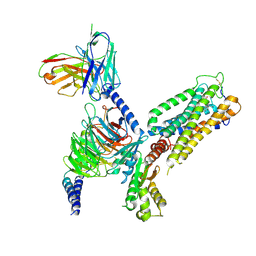 | | Cryo-EM structure of CB2-G protein complex | | Descriptor: | Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
7TKW
 
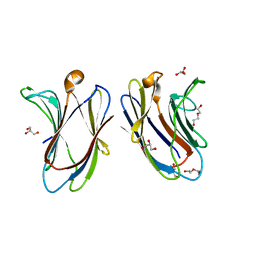 | |
7TKY
 
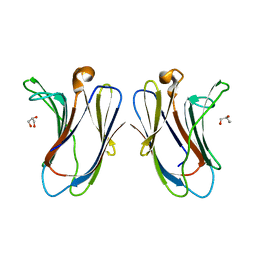 | |
7TKX
 
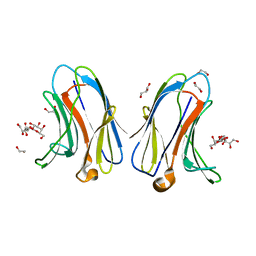 | | Crystal structure of R14A human Galectin-7 mutant in presence of 4-O-beta-D-Galactopyranosyl-D-glucose | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2,3,5, 6-tetrakis(oxidanyl)hexanal, 1,2-ETHANEDIOL, ... | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of R14A human Galectin-7 mutant in presence of 4-O-beta-D-Galactopyranosyl-D-glucose
To Be Published
|
|
7TKZ
 
 | |
7TRN
 
 | |
7TRO
 
 | | Crystal structure of R14A-R20A human Galectin-7 mutant in presence of lactose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galectin-7, ... | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-01-29 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of R14A-R20A human Galectin-7 mutant in presence of lactose
To Be Published
|
|
8GUT
 
 | | Cryo-EM structure of LEI-CB2-Gi complex | | Descriptor: | 1-[[4-[5-fluoranyl-6-[(oxan-4-ylamino)methyl]pyridin-2-yl]phenyl]methyl]-3-(2-methylpropyl)imidazolidine-2,4-dione, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Li, X.T, Chang, H, Wu, L.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8GUR
 
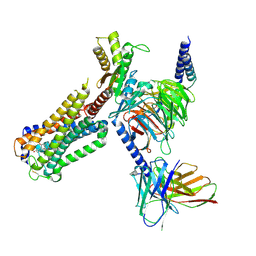 | | Cryo-EM structure of CP-CB2-G protein complex | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
8GFO
 
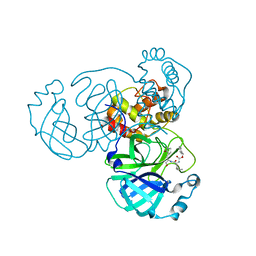 | |
8GFR
 
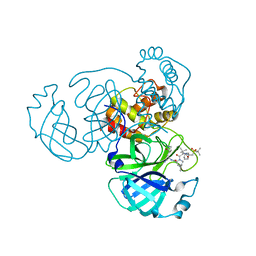 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with NBH2 | | Descriptor: | (1R,2S,5S)-N-{(1S)-1-cyano-2-[(3S)-2-oxopyrrolidin-3-yl]ethyl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
8GFU
 
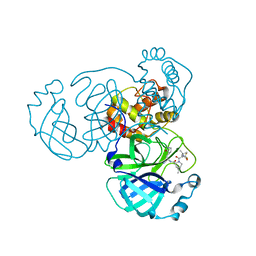 | |
8GFK
 
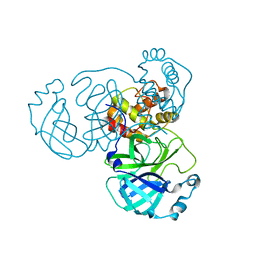 | |
5OSB
 
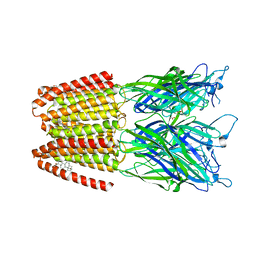 | | GLIC-GABAAR alpha1 chimera crystallized in complex with THDOC at pH4.5 | | Descriptor: | ACETATE ION, CHLORIDE ION, Proton-gated ion channel,Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Laverty, D.C, Gold, M.G, Smart, T.G. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8GFN
 
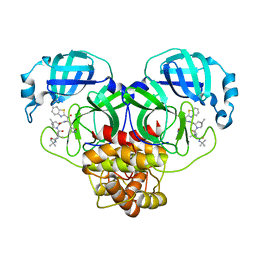 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with BBH1 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-(1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
3OK8
 
 | | I-BAR OF PinkBAR | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 2, GLYCEROL | | Authors: | Boczkowska, M, Rebowski, G, Saarikangas, J, Lappalainen, P, Dominguez, R. | | Deposit date: | 2010-08-24 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures.
Nat.Struct.Mol.Biol., 18, 2011
|
|
5OOM
 
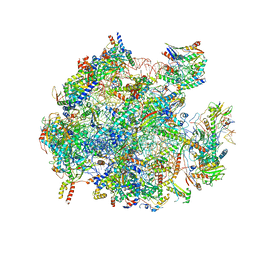 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8GIF
 
 | |
8GIE
 
 | |
