1POQ
 
 | | Solution Structure of a Superantigen from Yersinia pseudotuberculosis | | Descriptor: | YPM | | Authors: | Donadini, R, Liew, C.W, Kwan, A.H, Mackay, J.P, Fields, B.A. | | Deposit date: | 2003-06-16 | | Release date: | 2004-01-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Crystal and Solution Structures of a Superantigen from Yersinia pseudotuberculosis Reveal a Jelly-Roll Fold.
Structure, 12, 2004
|
|
1PP5
 
 | | Structure of Antibacterial Peptide Microcin J25: a 21-Residue Lariat Protoknot | | Descriptor: | microcin J25 | | Authors: | Bayro, M.J, Swapna, G.V.T, Huang, J.Y, Ma, L.-C, Mukhopadhyay, J, Ebright, R.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-16 | | Release date: | 2003-10-28 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Structure of Antibacterial Peptide Microcin J25: A 21-Residue Lariat Protoknot.
J.Am.Chem.Soc., 125, 2003
|
|
1PZ8
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
8BM7
 
 | |
1N9J
 
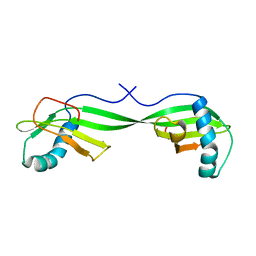 | | Solution Structure of the 3D domain swapped dimer of Stefin A | | Descriptor: | Cystatin A | | Authors: | Staniforth, R.A, Giannini, S, Higgins, L.D, Conroy, M.J, Hounslow, A.M, Jerala, R, Craven, C.J, Waltho, J.P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional domain swapping in the folded and molten-globule states of cystatins, an amyloid-forming structural superfamily
Embo J., 20, 2001
|
|
1PZ9
 
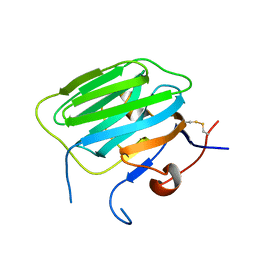 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1HY9
 
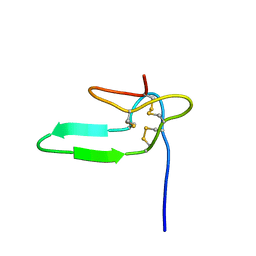 | | COCAINE AND AMPHETAMINE REGULATED TRANSCRIPT | | Descriptor: | COCAINE AND AMPHETAMINE REGULATED TRANSCRIPT PROTEIN | | Authors: | Ludvigsen, S, Thim, L, Blom, A.M, Wulff, B.S. | | Deposit date: | 2001-01-18 | | Release date: | 2001-08-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the satiety factor, CART, reveals new functionality of a well-known fold.
Biochemistry, 40, 2001
|
|
2AIT
 
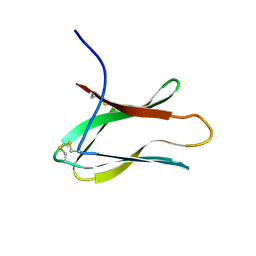 | | DETERMINATION OF THE COMPLETE THREE-DIMENSIONAL STRUCTURE OF THE ALPHA-AMYLASE INHIBITOR TENDAMISTAT IN AQUEOUS SOLUTION BY NUCLEAR MAGNETIC RESONANCE AND DISTANCE GEOMETRY | | Descriptor: | TENDAMISTAT | | Authors: | Kline, A.D, Braun, W, Guntert, P, Billeter, M, Wuthrich, K. | | Deposit date: | 1989-05-24 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the complete three-dimensional structure of the alpha-amylase inhibitor tendamistat in aqueous solution by nuclear magnetic resonance and distance geometry.
J.Mol.Biol., 204, 1988
|
|
2AWQ
 
 | |
2AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14Q | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-18 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1INZ
 
 | | SOLUTION STRUCTURE OF THE EPSIN N-TERMINAL HOMOLOGY (ENTH) DOMAIN OF HUMAN EPSIN | | Descriptor: | EPS15-INTERACTING PROTEIN(EPSIN) | | Authors: | Koshiba, S, Kigawa, T, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epsin N-terminal homology (ENTH) domain of human epsin
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
2ARW
 
 | | The solution structure of the membrane proximal cytokine receptor domain of the human interleukin-6 receptor | | Descriptor: | Interleukin-6 receptor alpha chain | | Authors: | Hecht, O, Dingley, A.J, Schwantner, A, Ozbek, S, Rose-John, S, Grotzinger, J. | | Deposit date: | 2005-08-22 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the membrane-proximal cytokine receptor domain of the human interleukin-6 receptor
Biol.Chem., 387, 2006
|
|
1IQ3
 
 | | SOLUTION STRUCTURE OF THE EPS15 HOMOLOGY DOMAIN OF A HUMAN POB1 | | Descriptor: | CALCIUM ION, RALBP1-INTERACTING PROTEIN (PARTNER OF RALBP1) | | Authors: | Koshiba, S, Kigawa, T, Iwahara, J, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-06 | | Release date: | 2001-06-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Eps15 homology domain of a human POB1 (partner of RalBP1).
FEBS Lett., 442, 1999
|
|
1KOT
 
 | |
1L8C
 
 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IXD
 
 | | Solution structure of the CAP-GLY domain from human cylindromatosis tomour-suppressor CYLD | | Descriptor: | Cylindromatosis tumour-suppressor CYLD | | Authors: | Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma
STRUCTURE, 12, 2004
|
|
1IYG
 
 | | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA | | Descriptor: | Hypothetical protein (2010003O14) | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Koshiba, S, Hamada, T, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-14 | | Release date: | 2003-02-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA
To be Published
|
|
1LX8
 
 | | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein | | Descriptor: | Excisionase | | Authors: | Sam, M.D, Papagiannis, C, Connolly, K.M, Corselli, L, Iwahara, J, Lee, J, Phillips, M, Wojciak, J.M, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2002-06-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein
J.Mol.Biol., 324, 2002
|
|
6O22
 
 | | Structure of Asf1-H3:H4-Rtt109-Vps75 histone chaperone-lysine acetyltransferase complex with the histone substrate. | | Descriptor: | Histone H3.2, Histone H4, Histone acetyltransferase RTT109, ... | | Authors: | Danilenko, N, Carlomagno, T, Kirkpatrick, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun, 10, 2019
|
|
6NHW
 
 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
2L29
 
 | | Complex structure of E4 mutant human IGF2R domain 11 bound to IGF-II | | Descriptor: | Insulin-like growth factor 2 receptor variant, Insulin-like growth factor II | | Authors: | Williams, C, Hoppe, H, Rezgui, D, Strickland, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Crump, M.P, Hassan, A.B. | | Deposit date: | 2010-08-13 | | Release date: | 2012-02-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
1J7O
 
 | |
6OSW
 
 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | Descriptor: | Forkhead box M1 | | Authors: | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
1B03
 
 | |
2N1K
 
 | |
