2OKI
 
 | | Crystal structure of dimeric form of PfFabZ in crystal form2 | | Descriptor: | Beta-hydroxyacyl-ACP dehydratase | | Authors: | Swarnamukhi, P.L, Sharma, S.K, Padala, P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Packing and loop-structure variations in non-isomorphous crystals of FabZ from Plasmodium falciparum
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2QOM
 
 | | The crystal structure of the E.coli EspP autotransporter Beta-domain. | | Descriptor: | Serine protease espP | | Authors: | Barnard, T.J, Dautin, N, Lukacik, P, Bernstein, H.D, Buchanan, S.K. | | Deposit date: | 2007-07-20 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Autotransporter structure reveals intra-barrel cleavage followed by conformational changes.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1FY8
 
 | | CRYSTAL STRUCTURE OF THE DELTAILE16VAL17 RAT ANIONIC TRYPSINOGEN-BPTI COMPLEX | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Ringe, D, Hedstrom, L. | | Deposit date: | 2000-09-28 | | Release date: | 2000-11-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
1I26
 
 | | SOLUTION STRUCTURE OF PTU-1, A TOXIN FROM THE ASSASSIN BUGS PEIRATES TURPIS THAT BLOCKS THE VOLTAGE SENSITIVE CALCIUM CHANNEL N-TYPE | | Descriptor: | PTU-1 | | Authors: | Bernard, C, Corzo, G, Mosbah, A, Nakajima, T, Darbon, H. | | Deposit date: | 2001-02-06 | | Release date: | 2001-11-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ptu1, a toxin from the assassin bug Peirates turpis that blocks the voltage-sensitive calcium channel N-type.
Biochemistry, 40, 2001
|
|
1EMX
 
 | | SOLUTION STRUCTURE OF HPTX2, A TOXIN FROM HETEROPODA VENATORIA SPIDER VENOM THAT BLOCKS KV4.2 POTASSIUM CHANNEL | | Descriptor: | HETEROPODATOXIN 2 | | Authors: | Bernard, C, Legros, C, Ferrat, G, Bishoff, U, Marquardt, A, Pongs, O, Darbon, H. | | Deposit date: | 2000-03-20 | | Release date: | 2001-01-24 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hpTX2, a toxin from Heteropoda venatoria spider that blocks Kv4.2 potassium channel.
Protein Sci., 9, 2000
|
|
4HU4
 
 | | Crystal structure of EAL domain of the E. coli DosP - dimeric form | | Descriptor: | Oxygen sensor protein DosP | | Authors: | Tarnawski, M, Barends, T.R.M, Hartmann, E, Schlichting, I. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the catalytic EAL domain of the Escherichia coli direct oxygen sensor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3FDO
 
 | |
1Q9G
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX
J.Mol.Biol., 336, 2004
|
|
4JAU
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4HU3
 
 | | Crystal structure of EAL domain of the E. coli DosP - monomeric form | | Descriptor: | Oxygen sensor protein DosP | | Authors: | Tarnawski, M, Barends, T.R.M, Hartmann, E, Schlichting, I. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structures of the catalytic EAL domain of the Escherichia coli direct oxygen sensor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JAS
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E and RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase, MAGNESIUM ION, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JA2
 
 | | Structural basis of a rationally rewired protein-protein interface (RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ACETATE ION, MAGNESIUM ION, Response regulator, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JAV
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853wt and RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Histidine kinase, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
1Q9F
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX.
J.Mol.Biol., 336, 2004
|
|
4JZY
 
 | | Crystal structures of Drosophila Cryptochrome | | Descriptor: | AMMONIUM ION, Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Czarna, A, Wolf, E. | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K09
 
 | |
4K06
 
 | | Crystal structure of MTX-II from Bothrops brazili venom complexed with polyethylene glycol | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MTX-II, ... | | Authors: | Fernandes, C.A.H, Comparetti, E.J, Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2013-04-03 | | Release date: | 2013-11-13 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural bases for a complete myotoxic mechanism: Crystal structures of two non-catalytic phospholipases A2-like from Bothrops brazili venom.
Biochim.Biophys.Acta, 1834, 2013
|
|
4K90
 
 | | Extracellular metalloproteinase from Aspergillus | | Descriptor: | BORIC ACID, CALCIUM ION, Extracellular metalloproteinase mep, ... | | Authors: | Fernandez, D, Russi, S, Vendrell, J, Monod, M, Pallares, I. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A functional and structural study of the major metalloprotease secreted by the pathogenic fungus Aspergillus fumigatus.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1K5W
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE SYNAPTOTAGMIN 1 C2B-DOMAIN: SYNAPTOTAGMIN 1 AS A PHOSPHOLIPID BINDING MACHINE | | Descriptor: | CALCIUM ION, Synaptotagmin I | | Authors: | Fernandez, I, Arac, D, Ubach, J, Gerber, S.H, Shin, O, Gao, Y, Anderson, R.G.W, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-10-12 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine.
Neuron, 32, 2001
|
|
1ORM
 
 | |
1KTX
 
 | | KALIOTOXIN (1-37) SHOWS STRUCTURAL DIFFERENCES WITH RELATED POTASSIUM CHANNEL BLOCKERS | | Descriptor: | KALIOTOXIN | | Authors: | Fernandez, I, Romi, R, Szendefi, S, Martin-Eauclaire, M.-F, Rochat, H, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1994-06-02 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Kaliotoxin (1-37) shows structural differences with related potassium channel blockers.
Biochemistry, 33, 1994
|
|
1LMR
 
 | | Solution of ADO1, a Toxin from the Assassin Bugs Agriosphodrus dohrni that Blocks the Voltage Sensitive Calcium Channel L-type | | Descriptor: | TOXIN ADO1 | | Authors: | Bernard, C, Corzo, G, Adachi-Akahane, S, Foures, G, Kanemaru, K, Furukawa, Y, Nakajima, T, Darbon, H. | | Deposit date: | 2002-05-02 | | Release date: | 2003-08-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ADO1, a toxin extracted from the saliva of the assassin bug, Agriosphodrus dohrni
Proteins: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3C0V
 
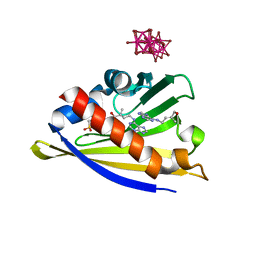 | | Crystal structure of cytokinin-specific binding protein in complex with cytokinin and Ta6Br12 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytokinin-specific binding protein, ... | | Authors: | Pasternak, O, Bujacz, A, Biesiadka, J, Bujacz, G, Sikorski, M, Jaskolski, M. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MAD phasing using the (Ta(6)Br(12))(2+) cluster: a retrospective study
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6W14
 
 | |
1XFQ
 
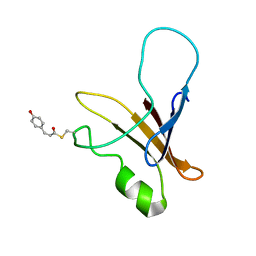 | | structure of the blue shifted intermediate state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
