8CHM
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfinyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfinyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CPB
 
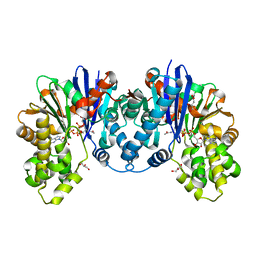 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
6QAZ
 
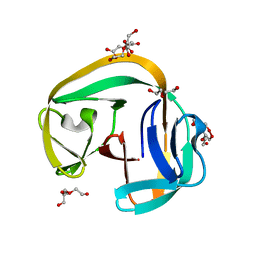 | | Crystal structure of gp41-1 intein | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Mikula, K.M, Beyer, H.M, Li, M, Wlodawer, A, Iwai, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the naturally split gp41-1 intein guides the engineering of orthogonal split inteins from cis-splicing inteins.
Febs J., 287, 2020
|
|
7P07
 
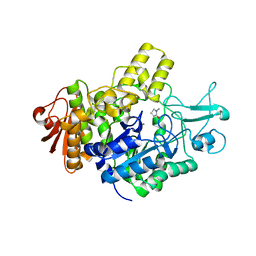 | | Structure of the maltase BaAG2 from Blastobotrys adeninivorans in complex with glucose | | Descriptor: | 1-methylpyrrolidin-2-one, BaAG2, CALCIUM ION, ... | | Authors: | Ernits, K, Visnapuu, T, Persson, K. | | Deposit date: | 2021-06-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Insight into a Yeast Maltase-The Ba AG2 from Blastobotrys adeninivorans with Transglycosylating Activity.
J Fungi (Basel), 7, 2021
|
|
8DCL
 
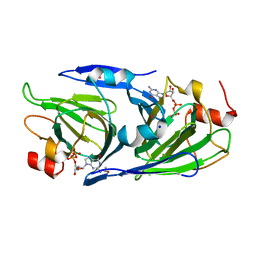 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase from campylobacter jejuni, serotype HS:23/36 | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
6NU8
 
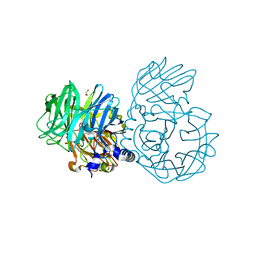 | |
7LVA
 
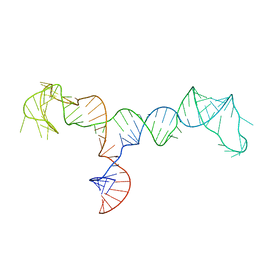 | | Solution structure of the HIV-1 PBS-segment | | Descriptor: | RNA (103-MER) | | Authors: | Heng, X, Song, Z. | | Deposit date: | 2021-02-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.
Nucleic Acids Res., 49, 2021
|
|
4X1T
 
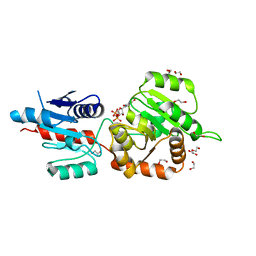 | | The crystal structure of Arabidopsis thaliana galactolipid synthase MGD1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Monogalactosyldiacylglycerol synthase 1, chloroplastic, ... | | Authors: | Rocha, J, Breton, C. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights and membrane binding properties of MGD1, the major galactolipid synthase in plants.
Plant J., 85, 2016
|
|
4X3J
 
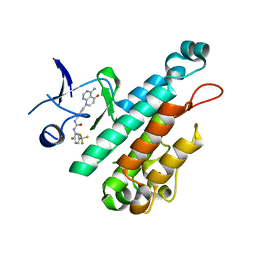 | | Selection of fragments for kinase inhibitor design: decoration is key | | Descriptor: | 1-[4-(4-amino-5-oxopyrido[2,3-d]pyrimidin-8(5H)-yl)phenyl]-3-[2-fluoro-5-(trifluoromethyl)phenyl]urea, Angiopoietin-1 receptor | | Authors: | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | Deposit date: | 2014-11-30 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
7P6I
 
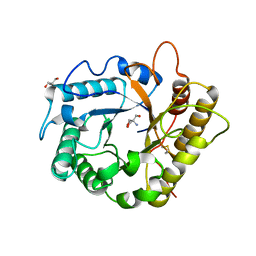 | | Crystal structure of the endoglucanase RBcel1 Y201F | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase | | Authors: | Collet, L, Dutoit, R. | | Deposit date: | 2021-07-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Highlighting the factors governing transglycosylation in the GH5_5 endo-1,4-beta-glucanase RBcel1.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7P7F
 
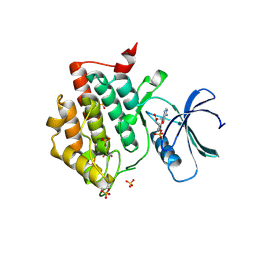 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
7ALU
 
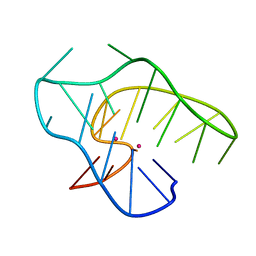 | |
4X1R
 
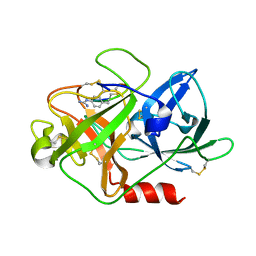 | | The crystal structure of mupain-1-12 in complex with murinised human uPA at pH7.4 | | Descriptor: | 1-phenylguanidine, Urokinase-type plasminogen activator, mupain-1-12 | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
6WZY
 
 | | Structure of DbNA(10) peptides bound to H-2Db MHC-I | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Farenc, C, Rossjohn, J. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Overlapping Peptides Elicit Distinct CD8 + T Cell Responses following Influenza A Virus Infection.
J Immunol., 205, 2020
|
|
4XPN
 
 | | Crystal Structure of Protein Phosphate 1 complexed with PP1 binding domain of GADD34 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Structural and Functional Analysis of the GADD34:PP1 eIF2 alpha Phosphatase.
Cell Rep, 11, 2015
|
|
7PPS
 
 | | apo FabB from Pseudomonas aeruginosa with single point mutation C161A | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 1, CHLORIDE ION, ... | | Authors: | Georgiou, C, Brenk, R, Yadrykhinsky, V. | | Deposit date: | 2021-09-15 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa FabB C161A, a template for structure-based design for new antibiotics.
F1000Res, 10, 2021
|
|
8ORK
 
 | | cyclic 2,3-diphosphoglycerate synthetase from the hyperthermophilic archaeon Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | De Rose, S.A, Isupov, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
Front Microbiol, 14, 2023
|
|
8DOM
 
 | |
7Q2H
 
 | | mycolic acid methyltransferase Hma (MmaA4) from Mycobac-terium tuberculosis in complex with ZT275 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Hydroxymycolate synthase MmaA4, ... | | Authors: | Maveyraud, L, Galy, R, Mourey, L. | | Deposit date: | 2021-10-25 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Ligand Discovery Applied to the Mycolic Acid Methyltransferase Hma (MmaA4) from Mycobacterium tuberculosis : A Crystallographic and Molecular Modelling Study.
Pharmaceuticals, 14, 2021
|
|
4XUC
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd18 (1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one) | | Descriptor: | 1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
7PUQ
 
 | | CARM1 in complex with EML982 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[3-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]propyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PPY
 
 | | CARM1 in complex with EML709 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
4XPP
 
 | | Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with D-galactose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase, beta-D-galactopyranose | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
7PV6
 
 | | CARM1 in complex with EML734 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, TETRAETHYLENE GLYCOL, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-10-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PPQ
 
 | | CARM1 in complex with EML736 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
