7LYQ
 
 | |
7LM8
 
 | |
7LQW
 
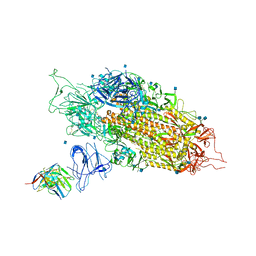 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
7LWW
 
 | |
7LWO
 
 | |
7LYM
 
 | |
7LWT
 
 | |
7M0J
 
 | |
7LWU
 
 | |
7LWV
 
 | |
7LWQ
 
 | |
7LWL
 
 | |
7LYL
 
 | |
7LJR
 
 | | SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab | | Descriptor: | Fab DH1043 heavy chain, Fab DH1043 light chain, Spike glycoprotein | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates.
Biorxiv, 2021
|
|
7LWS
 
 | |
7LWK
 
 | |
7LYO
 
 | |
7LWP
 
 | |
7LYP
 
 | |
7LYN
 
 | |
7LWN
 
 | |
7LXW
 
 | |
7LY0
 
 | |
7LXZ
 
 | |
7LY2
 
 | |
