3HBO
 
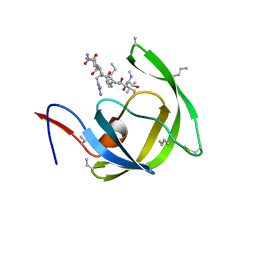 | | Crystal structure of chemically synthesized [D-Ala51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [D-Ala51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-04 | | Release date: | 2010-05-26 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1BAW
 
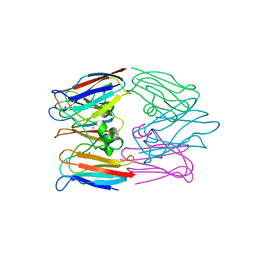 | | PLASTOCYANIN FROM PHORMIDIUM LAMINOSUM | | Descriptor: | COPPER (II) ION, PLASTOCYANIN, ZINC ION | | Authors: | Bond, C.S, Guss, J.M, Freeman, H.C, Wagner, M.J, Howe, C.J, Bendall, D.S. | | Deposit date: | 1998-04-19 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of plastocyanin from the cyanobacterium Phormidium laminosum.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2B4X
 
 | |
1G8V
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATF5UCGCG):5-FORMYLURIDINE/ ADENOSINE BASE-PAIRS IN B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*(UFR)P*CP*GP*CP*G)-3' | | Authors: | Tsunoda, M, Karino, N, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2000-11-21 | | Release date: | 2001-02-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallization and preliminary X-ray analysis of a DNA dodecamer containing 2'-deoxy-5-formyluridine; what is the role of magnesium cation in crystallization of Dickerson-type DNA dodecamers?
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2AIK
 
 | |
3RR3
 
 | | Structure of (R)-flurbiprofen bound to mCOX-2 | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Duggan, K.C, Hermanson, D.J, Musee, J, Prusakiewicz, J.J, Scheib, J, Carter, B.D, Banerjee, S, Oates, J.A, Marnett, L.J. | | Deposit date: | 2011-04-28 | | Release date: | 2011-11-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2.
Nat.Chem.Biol., 7, 2011
|
|
2AIJ
 
 | |
1Q47
 
 | | Structure of the Semaphorin 3A Receptor-Binding Module | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin 3A | | Authors: | Antipenko, A, Himanen, J.-P, van Leyen, K, Nardi-Dei, V, Lesniak, J, Barton, W.A, Rajashankar, K.R, Lu, M, Hoemme, C, Puschel, A, Nikolov, D. | | Deposit date: | 2003-08-01 | | Release date: | 2004-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the semaphorin-3A receptor binding module.
Neuron, 39, 2003
|
|
336D
 
 | | INTERACTION BETWEEN LEFT-HANDED Z-DNA AND POLYAMINE-3 THE CRYSTAL STRUCTURE OF THE D(CG)3 AND THERMOSPERMINE COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE | | Authors: | Ohishi, H, Terasoma, N, Nakanishi, I, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K.-I. | | Deposit date: | 1997-06-24 | | Release date: | 1998-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between left-handed Z-DNA and polyamine - 3. The crystal structure of the d(CG)3 and thermospermine complex.
FEBS Lett., 398, 1996
|
|
3AIS
 
 | | Crystal structure of a mutant beta-glucosidase in wheat complexed with DIMBOA-Glc | | Descriptor: | (2S)-2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one, Beta-glucosidase, beta-D-glucopyranose | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
4KC2
 
 | | Structure of the blood group glycosyltransferase AAglyB in complex with a pyridine inhibitor as a neutral pyrophosphate surrogate | | Descriptor: | 6-(1-beta-D-Galactopyranosyloxymethyl)-N-(5'-deoxyluridine-5'-yl)picolinamide, Fucosylglycoprotein alpha-N-acetylgalactosaminyltransferase soluble form, MANGANESE (II) ION, ... | | Authors: | Cuesta-Seijo, J.A, Wang, S, Lafont, D, Vidal, S, Palcic, M.M. | | Deposit date: | 2013-04-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of glycosyltransferase inhibitors: pyridine as a pyrophosphate surrogate.
Chemistry, 19, 2013
|
|
4CPE
 
 | | Wild-type streptavidin in complex with love-hate ligand 1 (LH1) | | Descriptor: | (3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N-{2-[(2,6- diphenylphenyl)formamido]ethyl}pentanamide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4BHV
 
 | | Measles virus phosphoprotein tetramerization domain | | Descriptor: | ACETATE ION, CALCIUM ION, PHOSPHOPROTEIN | | Authors: | Blocquel, D, Habchi, J, Durand, E, Sevajol, M, Ferron, F, Papageorgiou, N, Longhi, S. | | Deposit date: | 2013-04-08 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coiled-Coil Deformations in Crystal Structures: The Measles Virus Phosphoprotein Multimerization Domain as an Illustrative Example.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1MCQ
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1DTH
 
 | | METALLOPROTEASE | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, ATROLYSIN C, CALCIUM ION, ... | | Authors: | Botos, I, Scapozza, L, Zhang, D, Liotta, L.A, Meyer, E.F. | | Deposit date: | 1996-02-12 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Batimastat, a potent matrix mealloproteinase inhibitor, exhibits an unexpected mode of binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1MK6
 
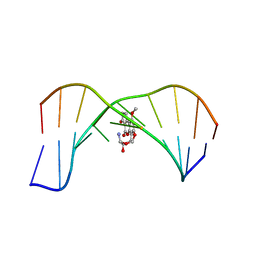 | | SOLUTION STRUCTURE OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT MISPAIRED WITH DEOXYADENOSINE | | Descriptor: | 5'-D(*AP*CP*AP*TP*CP*GP*AP*TP*CP*T)-3', 5'-D(*AP*GP*AP*TP*AP*GP*AP*TP*GP*T)-3', 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 | | Authors: | Giri, I, Johnston, D.S, Stone, M.P. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | MISPAIRING OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT WITH DEOXYADENOSINE RESULTS IN EXTRUSION OF THE MISMATCHED DA TOWARD THE MAJOR GROOVE
Biochemistry, 41, 2002
|
|
5RV5
 
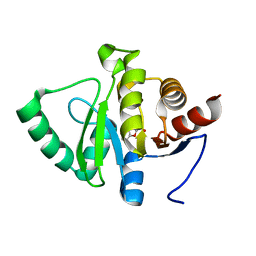 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000008578948 | | Descriptor: | BENZOFURO[3,2-D]PYRIMIDIN-4(3H)-ONE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3UKI
 
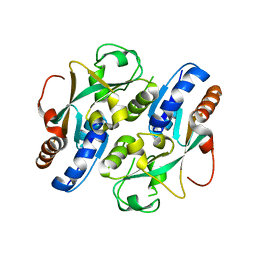 | | Crystal structure of reduced OxyR from Porphyromonas gingivalis | | Descriptor: | OxyR | | Authors: | Svintradze, D.V, Wright, H.T, Collazo-Santiago, E.A, Lewis, J.P. | | Deposit date: | 2011-11-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structures of the Porphyromonas gingivalis OxyR regulatory domain explain differences in expression of the OxyR regulon in Escherichia coli and P. gingivalis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1XCX
 
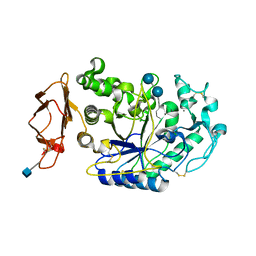 | | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Li, C, Begum, A, Numao, S, Park, K.H, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-03 | | Release date: | 2004-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts
Biochemistry, 44, 2005
|
|
2PII
 
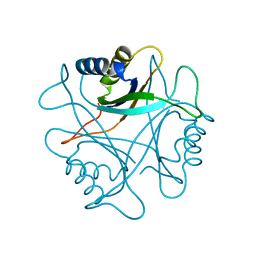 | | PII, GLNB PRODUCT | | Descriptor: | PII | | Authors: | Carr, P.D, Cheah, E, Suffolk, P.M, Ollis, D.L. | | Deposit date: | 1995-05-02 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the signal transduction protein from Escherichia coli at 1.9 A.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3D5O
 
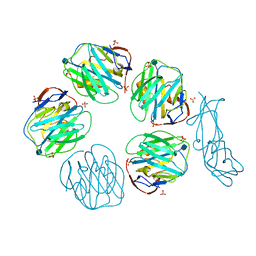 | | Structural recognition and functional activation of FcrR by innate pentraxins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low affinity immunoglobulin gamma Fc region receptor II-a, ... | | Authors: | Lu, J, Marnell, L.L, Marjon, K.D, Mold, C, Du Clos, T.W, Sun, P.D. | | Deposit date: | 2008-05-16 | | Release date: | 2008-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural recognition and functional activation of FcgammaR by innate pentraxins.
Nature, 456, 2008
|
|
2QKH
 
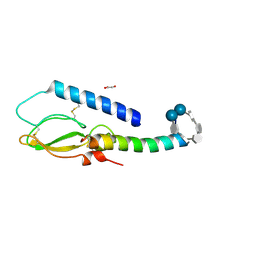 | | Crystal structure of the extracellular domain of human GIP receptor in complex with the hormone GIP | | Descriptor: | Cyclic 2,3-di-O-methyl-alpha-D-glucopyranose-(1-4)-2-O-methyl-alpha-D-glucopyranose-(1-4)-2,6-di-O-methyl-alpha-D-glucopyranose-(1-4)-2-O-methyl-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-3-O-methyl-alpha-D-glucopyranose, D(-)-TARTARIC ACID, Glucose-dependent insulinotropic polypeptide, ... | | Authors: | Parthier, C, Kleinschmidt, M, Neumann, P, Rudolph, R, Manhart, S, Schlenzig, D, Fanghanel, J, Rahfeld, J.-U, Demuth, H.-U, Stubbs, M.T. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5RSG
 
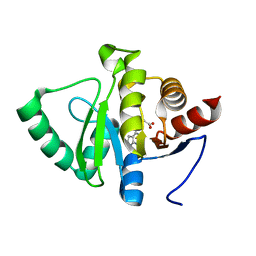 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000263392672 | | Descriptor: | N-methyl-N-7H-pyrrolo[2,3-d]pyrimidin-4-yl-beta-alanine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSH
 
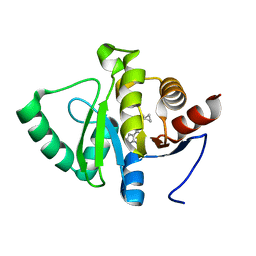 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000274438208 | | Descriptor: | 4-(5-azaspiro[2.5]octan-5-yl)-7H-pyrrolo[2,3-d]pyrimidine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVD
 
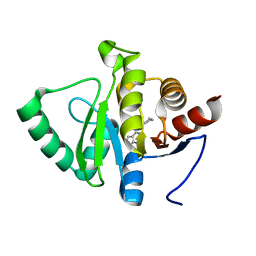 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000263980802 | | Descriptor: | 4-[(2R)-2-cyclobutylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
