9EN2
 
 | | Crystal structure of the metalloproteinase enhancer PCPE-1 complexed with nanobodies VHH-H4 and VHH-I5 | | Descriptor: | CALCIUM ION, GLYCEROL, Procollagen C-endopeptidase enhancer 1, ... | | Authors: | Lagoutte, P, Gueguen-Chaignon, V, Bourhis, J.-M, Vadon-Le Goff, S. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mono- and Bi-specific Nanobodies Targeting the CUB Domains of PCPE-1 Reduce the Proteolytic Processing of Fibrillar Procollagens.
J.Mol.Biol., 436, 2024
|
|
9EMV
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
9EMU
 
 | | RosC-8-demethyl-8-amino-FMN - Phosphate complex | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ermler, U, Mack, M, Demmer, U. | | Deposit date: | 2024-03-11 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Phosphatase RosC from Streptomyces davaonensis is Used for Roseoflavin Biosynthesis and has Evolved to Largely Prevent Dephosphorylation of the Important Cofactor Riboflavin-5'-phosphate.
J.Mol.Biol., 436, 2024
|
|
9EMS
 
 | | Crystal Structure of DC-SIGN in complex with JXH1902 | | Descriptor: | (2~{R},3~{S},4~{R},5~{S},6~{S})-6-[(2~{S})-3-azanyl-2-oxidanyl-propoxy]-2-(hydroxymethyl)-5-(4-phenyl-1,2,3-triazol-1-yl)oxane-3,4-diol, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Maier, T. | | Deposit date: | 2024-03-09 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thermodynamics-Guided Design Reveals a Cooperative Hydrogen Bond in DC-SIGN-targeted Glycomimetics.
J.Med.Chem., 67, 2024
|
|
9EMR
 
 | | Crystal Structure of DC-SIGN in complex with AL90 | | Descriptor: | (2~{R},3~{S},4~{R},5~{S},6~{S})-6-[(2~{S})-2,3-bis(oxidanyl)propoxy]-2-(hydroxymethyl)-5-(4-phenyl-1,2,3-triazol-1-yl)oxane-3,4-diol, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Maier, T. | | Deposit date: | 2024-03-09 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamics-Guided Design Reveals a Cooperative Hydrogen Bond in DC-SIGN-targeted Glycomimetics.
J.Med.Chem., 67, 2024
|
|
9EMQ
 
 | | Crystal Structure of DC-SIGN in complex with AL86 | | Descriptor: | (2~{R},3~{S},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-5-(4-phenyl-1,2,3-triazol-1-yl)-6-[[(3~{S})-piperidin-3-yl]methoxy]oxane-3,4-diol, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Maier, T. | | Deposit date: | 2024-03-09 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamics-Guided Design Reveals a Cooperative Hydrogen Bond in DC-SIGN-targeted Glycomimetics.
J.Med.Chem., 67, 2024
|
|
9EML
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
9EMK
 
 | | DupA from legionella covalently bound to ubiquitin-based probe | | Descriptor: | Polyubiquitin-B, Septation initiation protein, [(2~{R},3~{S},4~{R},5~{S})-5-[(1-ethyl-1,2,3-triazol-4-yl)methoxy]-3,4-bis(oxidanyl)oxolan-2-yl]methyl ethanesulfonate | | Authors: | Kim, R.Q, Kloet, M.S, van der Heden van Noort, G. | | Deposit date: | 2024-03-08 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Covalent Probes To Capture Legionella pneumophila Dup Effector Enzymes.
J.Am.Chem.Soc., 146, 2024
|
|
9EMJ
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with Toyocamycin and m7GpppA (Cap0-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EMI
 
 | |
9EME
 
 | | AL amyloid fibril from the FOR103 light chain | | Descriptor: | lambda 3 immunoglobulin light chain fragment, residues 2-116 | | Authors: | Pfeiffer, P.B, Karimi-Farsijani, S, Kupfer, N, Schmidt, M, Faendrich, M. | | Deposit date: | 2024-03-08 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Light chain mutations contribute to defining the fibril morphology in systemic AL amyloidosis.
Nat Commun, 15, 2024
|
|
9EMC
 
 | |
9EMB
 
 | | Structure of KefC Asp156Asn variant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE MONOPHOSPHATE, Glutathione-regulated potassium-efflux system protein KefC | | Authors: | Gulati, A, Kokane, S, Drew, D. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure of KefC Asp156Asn variant
To Be Published
|
|
9EMA
 
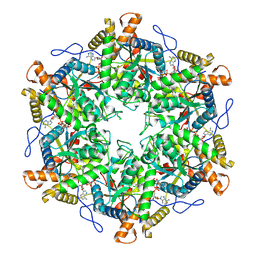 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
9EM9
 
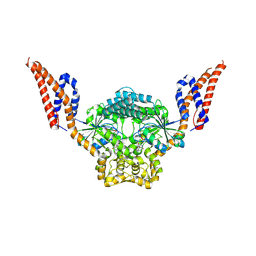 | | Structure of SynDLP MGD with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Slr0869 protein | | Authors: | Junglas, B, Gewehr, L, Schoennenbeck, P, Schneider, D, Sachse, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP.
Cell Rep, 43, 2024
|
|
9EM8
 
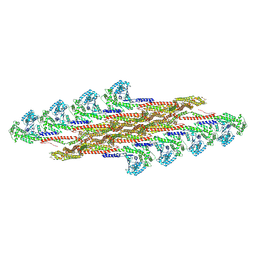 | | Oligomeric structure of SynDLP in presence of GDP | | Descriptor: | Slr0869 protein | | Authors: | Junglas, B, Gewehr, L, Schoennenbeck, P, Schneider, D, Sachse, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP.
Cell Rep, 43, 2024
|
|
9EM7
 
 | | Oligomeric structure of SynDLP in presence of GTP | | Descriptor: | Slr0869 protein | | Authors: | Junglas, B, Gewehr, L, Schoennenbeck, P, Schneider, D, Sachse, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP.
Cell Rep, 43, 2024
|
|
9EM6
 
 | |
9EM5
 
 | | OPR3 variant Y364P in its monomeric form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 12-oxophytodienoate reductase 3, CHLORIDE ION, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
9EM4
 
 | | OPR3 variant Y364P in its dimeric form obtained with ammonium sulfate | | Descriptor: | 12-oxophytodienoate reductase 3, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
9EM3
 
 | | OPR3 wild type in its monomeric form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
9EM2
 
 | |
9EM1
 
 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone and phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, Chronophin, GLYCEROL, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
9EM0
 
 | |
9E4V
 
 | | The structure of human vacuolar protein sorting 34 catalytic domain bound to MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Abiodun, W.O, Fullmer, A, Tsubaki, E, Doukov, T, Moody, J.D. | | Deposit date: | 2024-10-25 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Facile Bacterial Production of Human Vacuolar Protein Sorting 34 Enables Structural Characterization of Novel Inhibitors
To Be Published
|
|
