1DLJ
 
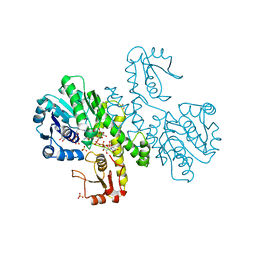 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
1DJ0
 
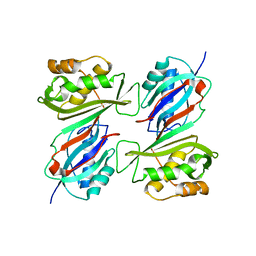 | | THE CRYSTAL STRUCTURE OF E. COLI PSEUDOURIDINE SYNTHASE I AT 1.5 ANGSTROM RESOLUTION | | Descriptor: | CHLORIDE ION, PSEUDOURIDINE SYNTHASE I | | Authors: | Foster, P.G, Huang, L, Santi, D.V, Stroud, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I.
Nat.Struct.Biol., 7, 2000
|
|
1DLI
 
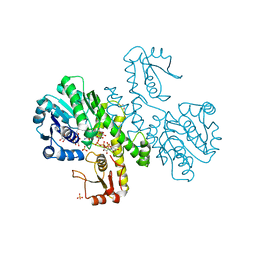 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
1ESY
 
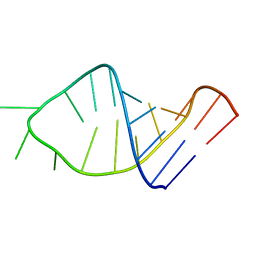 | |
1EVT
 
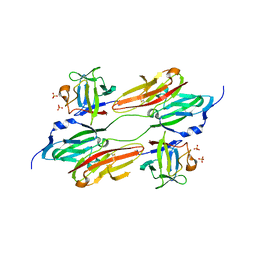 | | CRYSTAL STRUCTURE OF FGF1 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 1 (FGFR1) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 1), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 1), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1EWC
 
 | | CRYSTAL STRUCTURE OF ZN2+ LOADED STAPHYLOCOCCAL ENTEROTOXIN H | | Descriptor: | ENTEROTOXIN H, ZINC ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P, Antonsson, P, Svensson, A. | | Deposit date: | 2000-04-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
1EV2
 
 | | CRYSTAL STRUCTURE OF FGF2 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 2 (FGFR2) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 2), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 2), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1BZS
 
 | | CRYSTAL STRUCTURE OF MMP8 COMPLEXED WITH HMR2909 | | Descriptor: | 2-(BIPHENYL-4-SULFONYL)-1,2,3,4-TETRAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Schreuder, H, Brachvogel, V, Loenze, P. | | Deposit date: | 1998-11-04 | | Release date: | 2000-05-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Quantitative structure-activity relationship of human neutrophil collagenase (MMP-8) inhibitors using comparative molecular field analysis and X-ray structure analysis.
J.Med.Chem., 42, 1999
|
|
1EJ3
 
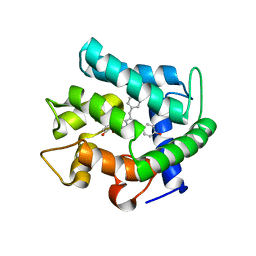 | | CRYSTAL STRUCTURE OF AEQUORIN | | Descriptor: | AEQUORIN, C2-HYDROPEROXY-COELENTERAZINE | | Authors: | Head, J.F, Inouye, S, Teranishi, K, Shimomura, O. | | Deposit date: | 2000-02-29 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the photoprotein aequorin at 2.3 A resolution.
Nature, 405, 2000
|
|
1QPE
 
 | |
1CNN
 
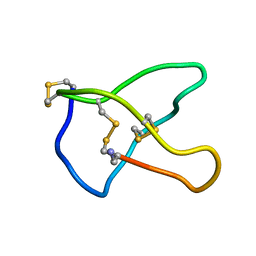 | | OMEGA-CONOTOXIN MVIIC FROM CONUS MAGUS | | Descriptor: | OMEGA-CONOTOXIN MVIIC | | Authors: | Nielsen, K.J, Adams, D, Thomas, L, Bond, T, Alewood, P.F, Craik, D.J, Lewis, R.J. | | Deposit date: | 1999-05-20 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of omega-conotoxins MVIIA, MVIIC and 14 loop splice hybrids at N and P/Q-type calcium channels.
J.Mol.Biol., 289, 1999
|
|
1QPJ
 
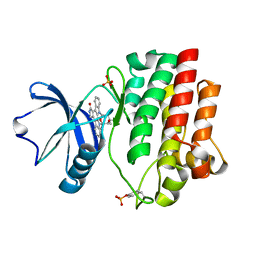 | | CRYSTAL STRUCTURE OF THE LYMPHOCYTE-SPECIFIC KINASE LCK IN COMPLEX WITH STAUROSPORINE. | | Descriptor: | LCK TYROSINE KINASE, STAUROSPORINE, SULFATE ION | | Authors: | Zhu, X, Kim, J.L, Rose, P.E, Stover, D.R, Toledo, L.M. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors.
Structure Fold.Des., 7, 1999
|
|
1QHR
 
 | | NOVEL COVALENT ACTIVE SITE THROMBIN INHIBITORS | | Descriptor: | 6-(2-HYDROXY-CYCLOPENTYL)-7-OXO-HEPTANAMIDINE, ALPHA THROMBIN, HIRUGEN | | Authors: | Jhoti, H, Cleasby, A, Reid, S, Thomas, P, Wonacott, A. | | Deposit date: | 1999-05-26 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thrombin complexed to a novel series of synthetic inhibitors containing a 5,5-trans-lactone template.
Biochemistry, 38, 1999
|
|
1DU2
 
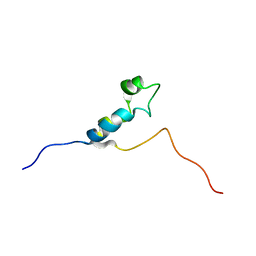 | | SOLUTION STRUCTURE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Descriptor: | DNA POLYMERASE III | | Authors: | Keniry, M.A, Berthon, H.A, Yang, J.-Y, Miles, C.S, Dixon, N.E. | | Deposit date: | 2000-01-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the theta subunit of DNA polymerase III from Escherichia coli.
Protein Sci., 9, 2000
|
|
1CJS
 
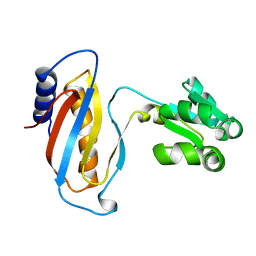 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 50S RIBOSOMAL PROTEIN L1P | | Authors: | Nevskaya, N, Tishchenko, S, Fedorov, R, Al-Karadaghi, S, Liljas, A, Kraft, A, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 1999-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family.
Structure Fold.Des., 8, 2000
|
|
1QHP
 
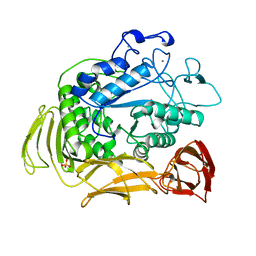 | | FIVE-DOMAIN ALPHA-AMYLASE FROM BACILLUS STEAROTHERMOPHILUS, MALTOSE COMPLEX | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Dauter, Z, Dauter, M, Brzozowski, A.M, Christensen, S, Borchert, T.V, Beier, L, Wilson, K.S, Davies, G.J. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of Novamyl, the five-domain "maltogenic" alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7A resolution.
Biochemistry, 38, 1999
|
|
1QHO
 
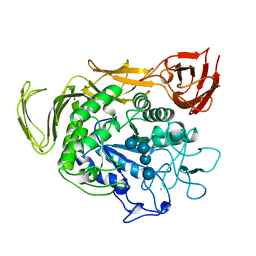 | | FIVE-DOMAIN ALPHA-AMYLASE FROM BACILLUS STEAROTHERMOPHILUS, MALTOSE/ACARBOSE COMPLEX | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Dauter, Z, Dauter, M, Brzozowski, A.M, Christensen, S, Borchert, T.V, Beier, L, Wilson, K.S, Davies, G.J. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of Novamyl, the five-domain "maltogenic" alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7A resolution.
Biochemistry, 38, 1999
|
|
1EIO
 
 | | ILEAL LIPID BINDING PROTEIN IN COMPLEX WITH GLYCOCHOLATE | | Descriptor: | GLYCOCHOLIC ACID, ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Hamilton, J.A, Sacchettini, J.C, Rueterjans, H. | | Deposit date: | 2000-02-27 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ileal lipid binding protein in complex with glycocholate.
Eur.J.Biochem., 267, 2000
|
|
3PFL
 
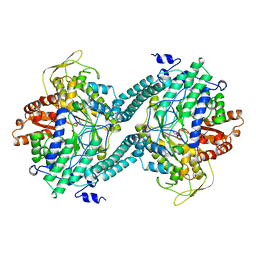 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI IN COMPLEX WITH SUBSTRATE ANALOGUE OXAMATE | | Descriptor: | OXAMIC ACID, PROTEIN (FORMATE ACETYLTRANSFERASE 1) | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1QHX
 
 | |
1QHS
 
 | |
1E04
 
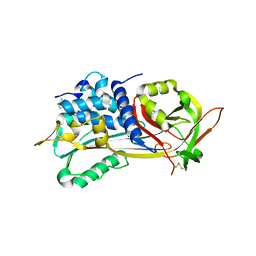 | | PLASMA BETA ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, GLYCEROL, ... | | Authors: | Mccoy, A.J, Skinner, R, Abrahams, J.-P, Pei, X.Y, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1E03
 
 | | PLASMA ALPHA ANTITHROMBIN-III AND PENTASACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, ... | | Authors: | McCoy, A.J, Jin, L, Abrahams, J.-P, Skinner, R, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1E05
 
 | | PLASMA ALPHA ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | McCoy, A.J, Skinner, R, Abrahams, J.-P, Pei, X.Y, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1E12
 
 | | Halorhodopsin, a light-driven chloride pump | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, HALORHODOPSIN, ... | | Authors: | Essen, L.-O, Kolbe, M, Oesterhelt, D. | | Deposit date: | 2000-04-14 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Light-Driven Chloride Pump Halorhodopsin at 1.8 A Resolution
Science, 288, 2000
|
|
