3OE7
 
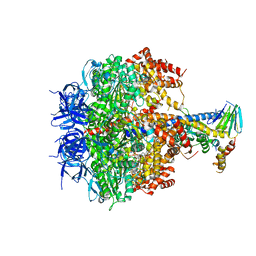 | | Structure of four mutant forms of yeast f1 ATPase: gamma-I270T | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
6U5T
 
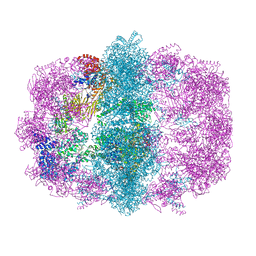 | |
8FGE
 
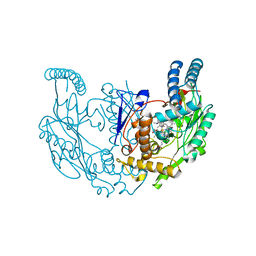 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with 4-(difluoromethyl)-6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)pyridin-2-amine dihydrochloride | | Descriptor: | 4-(difluoromethyl)-6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FGC
 
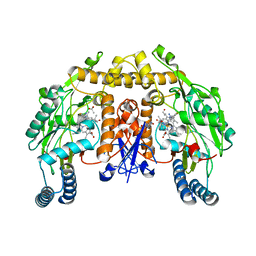 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(2-aminoethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{2-[5-(2-aminoethyl)-2,3-difluorophenyl]ethyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FGV
 
 | | Structure of rat neuronal nitric oxide synthase H692F mutant heme domain in complex with 6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-4-methoxypyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methoxypyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
3OFN
 
 | | Structure of four mutant forms of yeast F1 ATPase: alpha-N67I | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3X31
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 7,8-cis-14-epi-1a,25-Dihydroxy-19-norvitamin D3 | | Descriptor: | (1R,3R,7Z,14beta,17alpha)-17-[(2R)-6-hydroxy-6-methylheptan-2-yl]-9,10-secoestra-5,7-diene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2015-01-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Revisiting the 7,8-cis-vitamin D3 derivatives: synthesis, evaluating the biological activity, and study of the binding configuration
To be Published
|
|
8FGB
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-6-methylpyrimidin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FG9
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)pyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FGA
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(2-(dimethylamino)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(dimethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8947 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8FGD
 
 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with 6-(5-(2-(diethylamino)ethyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(diethylamino)ethyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 66, 2023
|
|
2LTQ
 
 | | High resolution structure of DsbB C41S by joint calculation with solid-state NMR and X-ray data | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Tang, M, Sperling, L.J, Schwieters, C.D, Nesbitt, A.E, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the Disulfide Bond Generating Membrane Protein DsbB in the Lipid Bilayer.
J.Mol.Biol., 425, 2013
|
|
3OPM
 
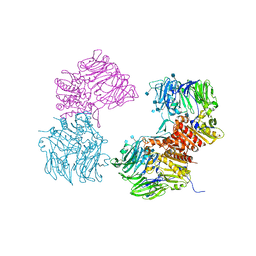 | | Crystal Structure of Human DPP4 Bound to TAK-294 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(aminomethyl)-2-(2-methylpropyl)-1-oxo-4-phenyl-1,2-dihydroisoquinolin-6-yl]oxy}acetamide, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-09-01 | | Release date: | 2011-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of 3-aminomethyl-1,2-dihydro-4-phenyl-1-isoquinolones: a new class of potent, selective, and orally active non-peptide dipeptidyl peptidase IV inhibitors that form a unique interaction with Lys554.
Bioorg.Med.Chem., 19, 2011
|
|
6U36
 
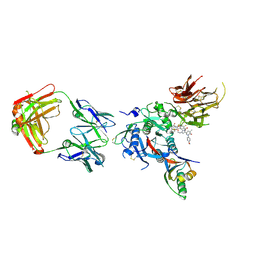 | | PCSK9 in complex with a Fab and compound 14 | | Descriptor: | 2-fluoro-4-{[(1R)-6-(2-{4-[1-(4-methoxyphenyl)-5-methyl-6-oxo-1,6-dihydropyridazin-3-yl]-1H-1,2,3-triazol-1-yl}ethoxy)-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-21 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
3X23
 
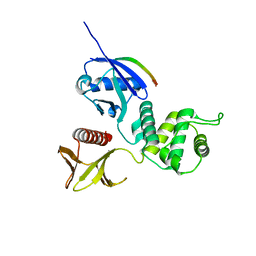 | | Radixin complex | | Descriptor: | Peptide from Matrix metalloproteinase-14, Radixin | | Authors: | Terawaki, S, Kitano, K, Aoyama, M, Mori, T, Hakoshima, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | MT1-MMP recognition by ERM proteins and its implication in CD44 shedding
Genes Cells, 20, 2015
|
|
2LQD
 
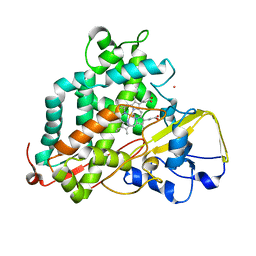 | | Reduced and CO-bound cytochrome P450cam (CYP101A1) | | Descriptor: | CARBON MONOXIDE, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Asciutto, E, Madura, J, Young, M.J. | | Deposit date: | 2012-03-02 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structural Ensembles of Substrate-Free Cytochrome P450(cam).
Biochemistry, 51, 2012
|
|
3P5W
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6U5U
 
 | |
6U9N
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 14 (TC-5139) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(4-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6UM8
 
 | | HIV Integrase in complex with Compound-14 | | Descriptor: | (2S)-tert-butoxy[7-(8-fluoro-5-methyl-3,4-dihydro-2H-1-benzopyran-6-yl)-5-methyl-2-phenylpyrazolo[1,5-a]pyrimidin-6-yl]acetic acid, DI(HYDROXYETHYL)ETHER, Integrase, ... | | Authors: | Khan, J.A, Kish, K. | | Deposit date: | 2019-10-09 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Optimization of Novel Pyrazolopyrimidines as Potent and Orally Bioavailable Allosteric HIV-1 Integrase Inhibitors.
J.Med.Chem., 63, 2020
|
|
3P5X
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3P5U
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3ZVG
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 98 | | Descriptor: | 3C PROTEASE, N-(tert-butoxycarbonyl)-O-tert-butyl-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
6U30
 
 | | The crystal structure of 4-pyridin-3-ylbenzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-3-yl)benzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
8FZ1
 
 | | Crystal structure of human PARP1 ART domain bound to inhibitor UKTT22 (compound 14) | | Descriptor: | (2P)-2-{3-[(2-amino-4,5-dimethylphenyl)carbamoyl]phenyl}-1H-benzimidazole-4-carboxamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel modifications of PARP inhibitor veliparib increase PARP1 binding to DNA breaks.
Biochem.J., 481, 2024
|
|
