4OOF
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203F mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
3GZN
 
 | | Structure of NEDD8-activating enzyme in complex with NEDD8 and MLN4924 | | Descriptor: | NEDD8, NEDD8-activating enzyme E1 catalytic subunit, NEDD8-activating enzyme E1 regulatory subunit, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2009-04-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ.
Mol.Cell, 37, 2010
|
|
6KKK
 
 | |
4F8P
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with galactose | | Descriptor: | ACETATE ION, TERT-BUTYL FORMATE, beta-D-galactopyranose, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1TIP
 
 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, PHOSPHOENZYME INTERMEDIATE OF FRU-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.-H, Olson, T.W, Ogata, C.M, Levitt, D.G, Banaszak, L.J, Lange, A.J. | | Deposit date: | 1997-05-28 | | Release date: | 1998-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a trapped phosphoenzyme during a catalytic reaction.
Nat.Struct.Biol., 4, 1997
|
|
3ZL1
 
 | | A thiazolyl-mannoside bound to FimH, monoclinic space group | | Descriptor: | CHLORIDE ION, N-{5-[(1R)-1-hydroxyethyl]-1,3-thiazol-2-yl}-alpha-D-mannopyranosylamine, PROTEIN FIMH | | Authors: | Brument, S, Sivignon, A, Dumych, T.I, Moreau, N, Roos, G, Guerardel, Y, Chalopin, T, Deniaud, D, Bilyy, R.O, Darfeuille-Michaud, A, Bouckaert, J, Gouin, S.G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Thiazolylaminomannosides as Potent Antiadhesives of Type 1 Piliated Escherichia Coli Isolated from Crohn'S Disease Patients.
J.Med.Chem., 56, 2013
|
|
3PZI
 
 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose | | Descriptor: | Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5, beta-D-glucopyranose | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3GJK
 
 | | crystal structure of a DNA duplex containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3', POTASSIUM ION | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S, Haraguchi, T, Xiao, M, Kurose, T, Ohkubo, A, Sekine, M, Shibata, T, Millington, C.L, Williams, D.M. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the stabilizing contributions of bicyclic cytosine analogues: crystal structures of DNA duplexes containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one
To be Published
|
|
1IER
 
 | | CUBIC CRYSTAL STRUCTURE OF NATIVE HORSE SPLEEN FERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Granier, T, Gallois, B, Dautant, A, Langlois D'Estaintot, B, Precigoux, G. | | Deposit date: | 1996-05-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparison of the structures of the cubic and tetragonal forms of horse-spleen apoferritin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
4L7G
 
 | | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxy-benzylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against Beta-Secretase-1 (BACE1) | | Descriptor: | (3aS,7aR)-7a-[3-(pyrimidin-5-yl)phenyl]-3a,6,7,7a-tetrahydro-4H-pyrano[4,3-d][1,3]oxazol-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE | | Authors: | Huestis, M.P, Liu, W, Volgraf, M, Purkey, H, Wang, W, Yu, C, Wu, P, Smith, D, Vigers, G, Dutcher, D, Geck Do, M.K, Hunt, K.W, Siu, M. | | Deposit date: | 2013-06-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxycycloalkylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against β-Secretase-1 (BACE-1)
Tetrahedron Lett., 2013
|
|
1FCY
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARBETA/GAMMA-SELECTIVE RETINOID CD564 | | Descriptor: | 6-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALENE-2-CARBONYL)-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1F7N
 
 | | CRYSTAL STRUCTURES OF FELINE IMMUNODEFICIENCY VIRUS DUTP PYROPHOSPHATASE AND ITS NUCLEOTIDE COMPLEXES IN THREE CRYSTAL FORMS. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, MAGNESIUM ION, POL POLYPROTEIN | | Authors: | Prasad, G.S, Stura, E.A, Elder, J.H, Stout, C.D. | | Deposit date: | 2000-06-27 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of feline immunodeficiency virus dUTP pyrophosphatase and its nucleotide complexes in three crystal forms.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3LJ8
 
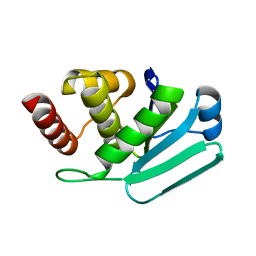 | | Crystal Structure of MKP-4 | | Descriptor: | Tyrosine-protein phosphatase | | Authors: | Jeong, D.G, Yoon, T.S, Jung, S.-K, Park, H.S, Ryu, S.E, Kim, S.J. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring binding sites other than the catalytic core in the crystal structure of the catalytic domain of MKP-4
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1F7U
 
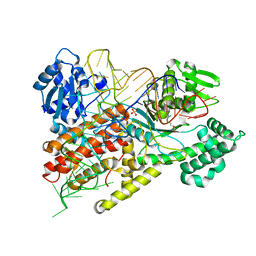 | | CRYSTAL STRUCTURE OF THE ARGINYL-TRNA SYNTHETASE COMPLEXED WITH THE TRNA(ARG) AND L-ARG | | Descriptor: | ARGININE, ARGINYL-TRNA SYNTHETASE, SULFATE ION, ... | | Authors: | Delagoutte, B, Moras, D, Cavarelli, J. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding
EMBO J., 19, 2000
|
|
1IMR
 
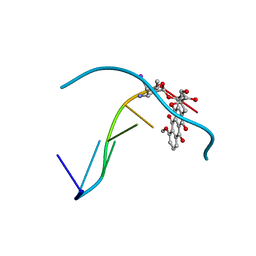 | | MOLECULAR STRUCTURE OF THE HALOGENATED ANTI-CANCER DRUG IODODOXORUBICIN COMPLEXED WITH D(TGTACA) AND D(CGATCG) | | Descriptor: | 4'-DEOXY-4'-IODODOXORUBICIN, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3') | | Authors: | Berger, I, Su, L, Spitzner, J.R, Kang, C, Burke, T.G, Rich, A. | | Deposit date: | 1995-10-23 | | Release date: | 1996-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular structure of the halogenated anti-cancer drug iododoxorubicin complexed with d(TGTACA) and d(CGATCG).
Nucleic Acids Res., 23, 1995
|
|
7ZGI
 
 | | chloroplast trigger factor (TIG1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidylprolyl isomerase, SULFATE ION | | Authors: | Carius, Y, Ries, F, Gries, K, Trentmann, O, Willmund, F, Lancaster, C.R.D. | | Deposit date: | 2022-04-03 | | Release date: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural features of chloroplast trigger factor determined at 2.6 angstrom resolution.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4CC1
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4C4S
 
 | |
3OKE
 
 | | Crystal structure of S25-39 in complex with Ko | | Descriptor: | S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ZINC ION, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-24 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
2ZBL
 
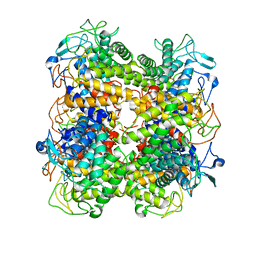 | | Functional annotation of Salmonella enterica yihS-encoded protein | | Descriptor: | Putative isomerase, beta-D-mannopyranose | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
4KQ1
 
 | |
4F8L
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with galactose and AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, BROMIDE ION, GLYCINE, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1RNB
 
 | |
1TI8
 
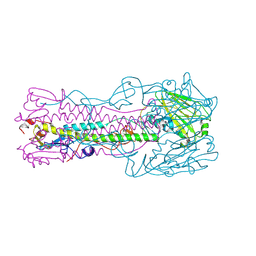 | | H7 Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose, ... | | Authors: | Russell, R.J, Gamblin, S.J, Haire, L.F, Stevens, D.J, Xaio, B, Ha, Y, Skehel, J.J. | | Deposit date: | 2004-06-02 | | Release date: | 2005-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes.
Virology, 325, 2004
|
|
2Z75
 
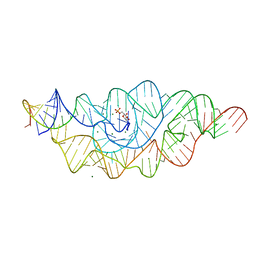 | | T. tengcongensis glmS ribozyme bound to glucosamine-6-phosphate | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, glmS ribozyme RNA, ... | | Authors: | Klein, D.J, Wilkinson, S.R, Been, M.D, Ferre-D'Amare, A.R. | | Deposit date: | 2007-08-15 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme
J.Mol.Biol., 373, 2007
|
|
