8I34
 
 | | The crystal structure of EPD-BCP1 from a marine sponge | | Descriptor: | (2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E},16~{E})-4,8,13,17-tetramethyl-3-oxidanyl-19-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]-1-[(1~{R},4~{S})-1,2,2-trimethyl-4-oxidanyl-cyclopentyl]nonadeca-2,4,6,8,10,12,14,16-octaen-18-yn-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASTAXANTHIN, ... | | Authors: | Shomura, Y, Kawasaki, S. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | An ependymin-related blue carotenoprotein decorates marine blue sponge.
J.Biol.Chem., 299, 2023
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
3RZN
 
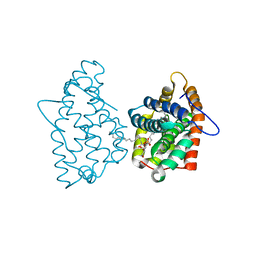 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
8G5U
 
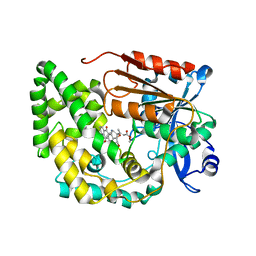 | | Crystal structure of TnmK2 complexed with TNM B | | Descriptor: | TnmK2, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2023-02-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis.
Nat.Chem.Biol., 20, 2024
|
|
4Z22
 
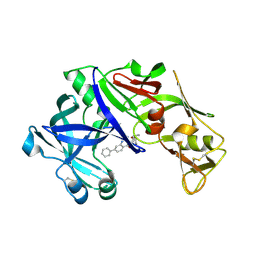 | | structure of plasmepsin II from Plasmodium Falciparum complexed with inhibitor DR718A | | Descriptor: | 2-amino-7-phenyl-3-{[(2R,5S)-5-phenyltetrahydrofuran-2-yl]methyl}quinazolin-4(3H)-one, Plasmepsin-2 | | Authors: | Recacha, R, Leitans, J, Tars, K, Jaudzems, K. | | Deposit date: | 2015-03-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Fragment-Based Discovery of 2-Aminoquinazolin-4(3H)-ones As Novel Class Nonpeptidomimetic Inhibitors of the Plasmepsins I, II, and IV.
J.Med.Chem., 59, 2016
|
|
5M81
 
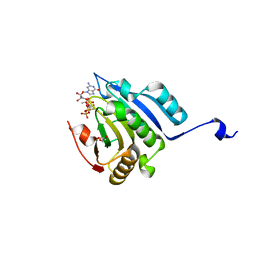 | | Translation initiation factor 4E in complex with (SP)-iPr-m7GppSpG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(3~{a}~{R},4~{R},6~{R},6~{a}~{R})-4-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-2,2-dimethyl-3~{a},4,6,6~{a}-tetrahydrofuro[3,4-d][1,3]dioxol-6-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-iPr-m7GppSpG mRNA 5' cap analog
To Be Published
|
|
4Z35
 
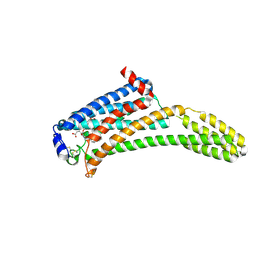 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
7O9W
 
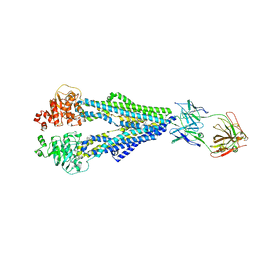 | | Encequidar-bound human P-glycoprotein in complex with UIC2-Fab | | Descriptor: | Multidrug resistance protein 1, UIC2 Fab-fragment heavy chain, UIC2 Fab-fragment light chain, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-04-17 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Discovery and Characterization of Potent Dual P-Glycoprotein and CYP3A4 Inhibitors: Design, Synthesis, Cryo-EM Analysis, and Biological Evaluations.
J.Med.Chem., 65, 2022
|
|
7NRB
 
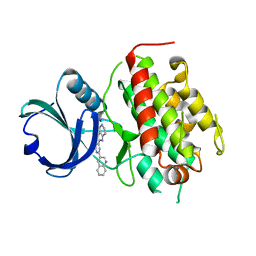 | | Re-refinement of MK3-inhibitor complex | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 3 | | Authors: | Croll, T.I, Read, R.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adaptive Cartesian and torsional restraints for interactive model rebuilding.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6C6E
 
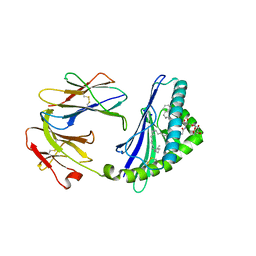 | | Structure of glycolipid aGSA[26,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
4IN7
 
 | | (M)L214N mutant of the Rhodobacter sphaeroides Reaction Center | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Saer, R.G, Hardjasa, A, Murphy, M.E.P, Beatty, J.T. | | Deposit date: | 2013-01-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Role of Rhodobacter sphaeroides photosynthetic reaction center residue M214 in the composition, absorbance properties, and conformations of H(A) and B(A) cofactors.
Biochemistry, 52, 2013
|
|
8IJC
 
 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5MUT
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 2-methyl-5-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,3-triazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
5MEV
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
7TI0
 
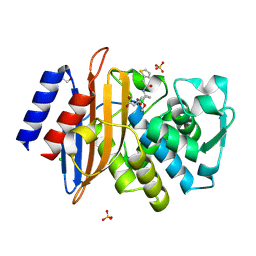 | | Structure of CTX-M-15 bound to RPX-7063 at 1.5A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Abendroth, J, Hecker, S.J, Edwards, T.E. | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
5MIO
 
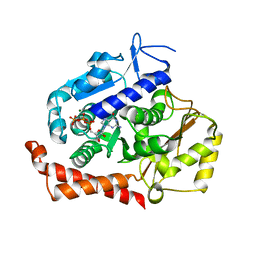 | | KIF2C-DARPIN FUSION PROTEIN BOUND TO TUBULIN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF2C,KIF2C FUSED TO A DARPIN,KIF2C FUSED TO A DARPIN, ... | | Authors: | Wang, W, Gigant, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin.
Nat Commun, 8, 2017
|
|
4ZAY
 
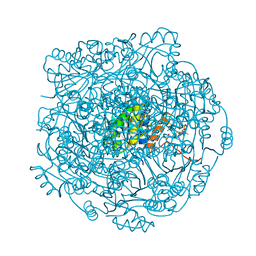 | | Structure of UbiX E49Q in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
6Z51
 
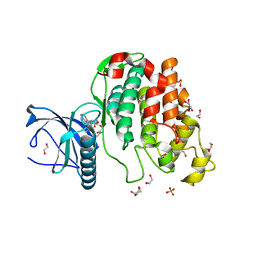 | | Crystal structure of CLK3 in complex with macrocycle ODS2002941 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, N-cyclopentyl-2-[(11,15-dimethyl-10-oxo-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-6-yl)oxy]acetamide, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2002941
To Be Published
|
|
5ZCP
 
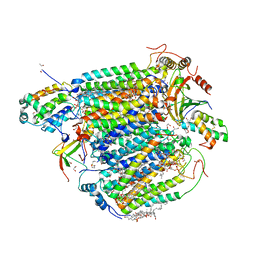 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 20 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
6Z5A
 
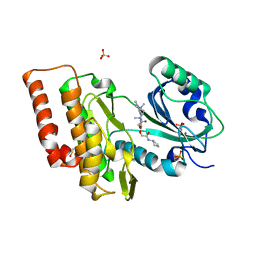 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941 | | Descriptor: | DIMETHYL SULFOXIDE, N-cyclopentyl-2-[(11,15-dimethyl-10-oxo-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-6-yl)oxy]acetamide, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941
To Be Published
|
|
7NRY
 
 | | Re-refinement of MAPKAP kinase-2/inhibitor complex 3fyj | | Descriptor: | (10R)-10-methyl-3-(6-methylpyridin-3-yl)-9,10,11,12-tetrahydro-8H-[1,4]diazepino[5',6':4,5]thieno[3,2-f]quinolin-8-one, CHLORIDE ION, MALONIC ACID, ... | | Authors: | Croll, T.I, Read, R.J. | | Deposit date: | 2021-03-04 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Adaptive Cartesian and torsional restraints for interactive model rebuilding.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7TI1
 
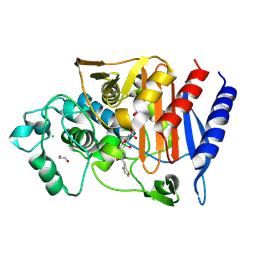 | | Structure of AmpC bound to RPX-7063 at 2.0A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Abendroth, J, Edwards, T.E, Hecker, S.J. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
6GCC
 
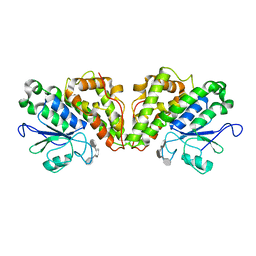 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor in complex with dextran-sulfate | | Descriptor: | 3,4-di-O-sulfo-alpha-D-glucopyranose-(1-6)-3,4-di-O-sulfo-alpha-D-altropyranose-(1-6)-1,2,3,4-tetra-O-sulfo-alpha-D-allopyranose, Glutathione transferase Xi 3 mutant C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-04-17 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
5ZGH
 
 | | Cryo-EM structure of the red algal PSI-LHCR | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pi, X. | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5N0E
 
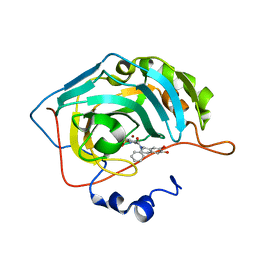 | | Crystal structure of human carbonic anhydrase II in complex with (S)-4-(6,7-dihydroxy-1-phenyl-3,4-tetrahydroisoquinoline-1H-2-carbonyl)benzenesulfonamide. | | Descriptor: | 4-[[(1~{S})-6,7-bis(oxidanyl)-1-phenyl-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing Molecular Interactions between Human Carbonic Anhydrases (hCAs) and a Novel Class of Benzenesulfonamides.
J. Med. Chem., 60, 2017
|
|
