6VTD
 
 | | Naegleria gruberi RNA ligase R149A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTF
 
 | | Naegleria gruberi RNA ligase with PPi | | Descriptor: | PYROPHOSPHATE 2-, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTB
 
 | | Naegleria gruberi RNA ligase K326A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT1
 
 | | Naegleria gruberi RNA ligase D172A mutant apo | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT8
 
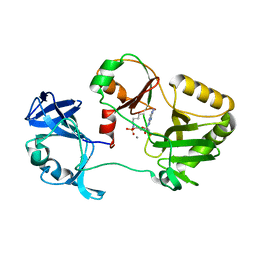 | | Naegleria gruberi RNA ligase E312A mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
4NJC
 
 | |
2F3I
 
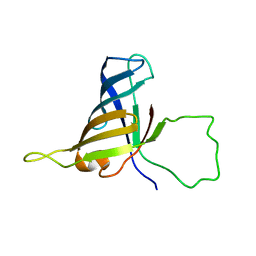 | | Solution Structure of a Subunit of RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerases I, II, and III 17.1 kDa polypeptide | | Authors: | Kang, X, Jin, C. | | Deposit date: | 2005-11-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural, biochemical, and dynamic characterizations of the hRPB8 subunit of human RNA polymerases
J.Biol.Chem., 281, 2006
|
|
6E8T
 
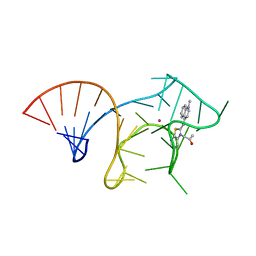 | |
6E8U
 
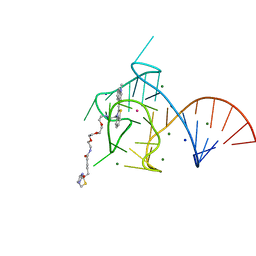 | | Structure of the Mango-III (A10U) aptamer bound to TO1-Biotin | | Descriptor: | 4-[(3-{2,16-dioxo-20-[(3aR,4R,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.
Nat. Chem. Biol., 15, 2019
|
|
1B42
 
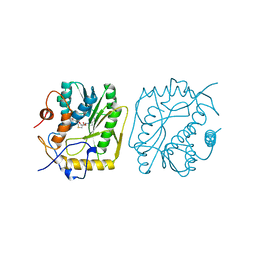 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M1ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-1-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4KYY
 
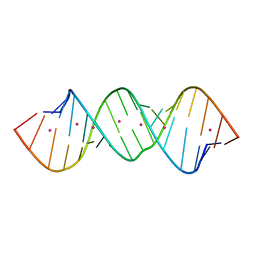 | |
1NA2
 
 | |
3ULV
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.522 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3ULU
 
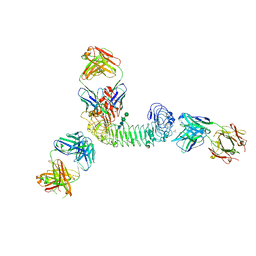 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
1G5H
 
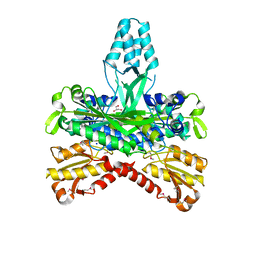 | | CRYSTAL STRUCTURE OF THE ACCESSORY SUBUNIT OF MURINE MITOCHONDRIAL POLYMERASE GAMMA | | Descriptor: | GLYCEROL, MITOCHONDRIAL DNA POLYMERASE ACCESSORY SUBUNIT, SODIUM ION | | Authors: | Carrodeguas, J.A, Theis, K, Bogenhagen, D.F, Kisker, C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer.
Mol.Cell, 7, 2001
|
|
1G5I
 
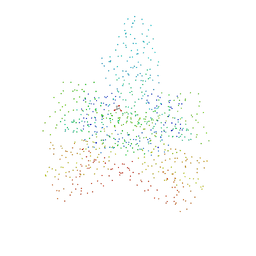 | | CRYSTAL STRUCTURE OF THE ACCESSORY SUBUNIT OF MURINE MITOCHONDRIAL POLYMERASE GAMMA | | Descriptor: | GLYCEROL, MITOCHONDRIAL DNA POLYMERASE ACCESSORY SUBUNIT, SODIUM ION | | Authors: | Carrodeguas, J.A, Theis, K, Bogenhagen, D.F, Kisker, C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer.
Mol.Cell, 7, 2001
|
|
2GVO
 
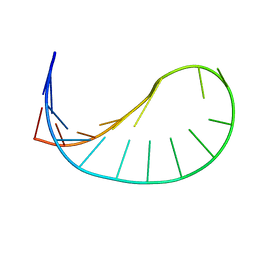 | | Solution structure of a purine rich hexaloop hairpin belonging to PGY/MDR1 mRNA and targeted by antisense oligonucleotides | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Joli, F, Bouchemal, N, Laigle, A, Hartmann, B, Hantz, E. | | Deposit date: | 2006-05-03 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a purine rich hexaloop hairpin belonging to PGY/MDR1 mRNA and targeted by antisense oligonucleotides.
Nucleic Acids Res., 34, 2006
|
|
1CKO
 
 | |
6ZMC
 
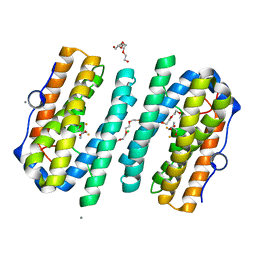 | | Structure of the tRNA-Monooxygenase enzyme MiaE frozen under 2000 bar using the high pressure freezing method | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGON, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
6ZMB
 
 | | Structure of the native tRNA-Monooxygenase enzyme MiaE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
6ZMA
 
 | | Structure of the tRNA-Monooxygenase enzyme MiaE frozen under 140 bar of krypton using the soak and freeze methodology | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
2OKO
 
 | | Z. mobilis tRNA guanine transglycosylase E235Q mutant apo-structure at pH 5.5 | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N. | | Deposit date: | 2007-01-17 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Glutamate versus Glutamine Exchange Swaps Substrate Selectivity in tRNA-Guanine Transglycosylase: Insight into the Regulation of Substrate Selectivity by Kinetic and Crystallographic Studies.
J.Mol.Biol., 374, 2007
|
|
2QII
 
 | | Crystal Structure Of tRNA-Guanine Transglycosylase (TGT) From Zymomonas mobilis Complexed With Archaeosine Precursor, Preq0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Brenk, R, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-07-04 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glutamate versus glutamine exchange swaps substrate selectivity in tRNA-guanine transglycosylase: insight into the regulation of substrate selectivity by kinetic and crystallographic studies.
J.Mol.Biol., 374, 2007
|
|
2QZV
 
 | | Draft Crystal Structure of the Vault Shell at 9 Angstroms Resolution | | Descriptor: | Major vault protein | | Authors: | Anderson, D.H, Kickhoefer, V.A, Sievers, S.A, Rome, L.H, Eisenberg, D. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Draft crystal structure of the vault shell at 9-A resolution.
Plos Biol., 5, 2007
|
|
3FAR
 
 | |
