7AXX
 
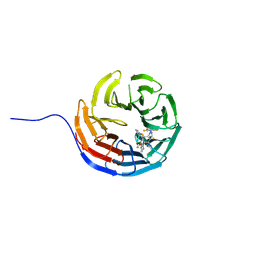 | |
7AXS
 
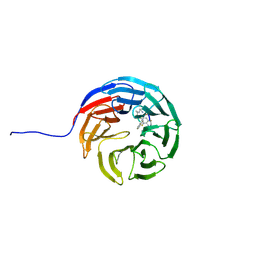 | |
4GMR
 
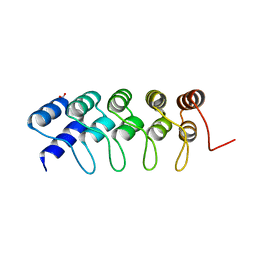 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR266. | | Descriptor: | NITRATE ION, OR266 DE NOVO PROTEIN | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Mao, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
7B69
 
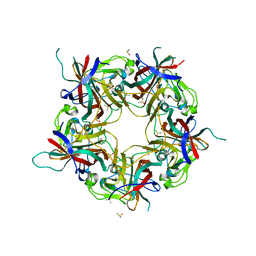 | |
7B6A
 
 | | BK Polyomavirus VP1 pentamer core (residues 30-299) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Osipov, E.M, Munawar, A, Beelen, S, Strelkov, S.V. | | Deposit date: | 2020-12-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
7B6C
 
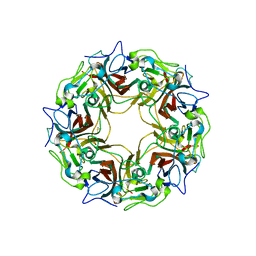 | | BK Polyomavirus VP1 pentamer fusion with long C-terminal extended arm | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Major capsid protein VP1,Major capsid protein VP1 | | Authors: | Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2020-12-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
6ZM3
 
 | | The structure of an E2 ubiquitin-conjugating complex (UBC2-UEV1) essential for Leishmania amastigote differentiation | | Descriptor: | Putative ubiquitin-conjugating enzyme e2, Ubiquitin-conjugating enzyme-like protein | | Authors: | Burge, R.J, Dodson, E.J, Wilkinson, A.J, Mottram, J.C. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leishmania differentiation requires ubiquitin conjugation mediated by a UBC2-UEV1 E2 complex.
Plos Pathog., 16, 2020
|
|
6ZPO
 
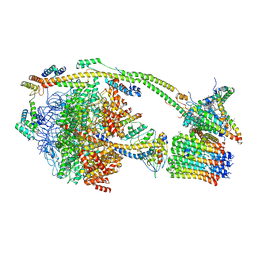 | | bovine ATP synthase monomer state 1 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZTJ
 
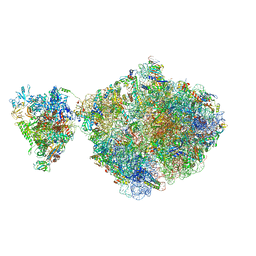 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZVI
 
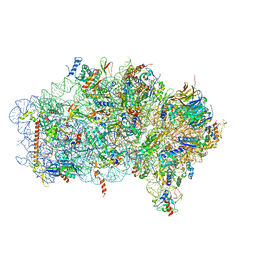 | | Mbf1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
7BVB
 
 | |
6ZU5
 
 | | Structure of the Paranosema locustae ribosome in complex with Lso2 | | Descriptor: | 18S rRNA, 25S rRNA, 5S rRNA, ... | | Authors: | Ehrenbolger, K, Jespersen, N, Sharma, H, Sokolova, Y.Y, Tokarev, Y.S, Vossbrinck, C.R, Barandun, J. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Differences in structure and hibernation mechanism highlight diversification of the microsporidian ribosome.
Plos Biol., 18, 2020
|
|
1I7Y
 
 | |
6ZTN
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
5YF5
 
 | |
6ZQM
 
 | | bovine ATP synthase monomer state 2 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-10 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZTM
 
 | | E. coli 70S-RNAP expressome complex in collided state without NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZVH
 
 | | EDF1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
8UTX
 
 | | Solution structure of a 12-mer peptide bearing a bicyclic Asx motif mimic (BAMM) as a synthetic N-cap | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, TRP-CYS-ASP-ALA-ALA-CYS-CYS-ALA-ALA-ALA-LYS-ALA-NH2 peptide | | Authors: | Mi, T.X, Burgess, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Bioinformatics leading to conveniently accessible, helix enforcing, bicyclic ASX motif mimics (BAMMs).
Nat Commun, 15, 2024
|
|
7BXU
 
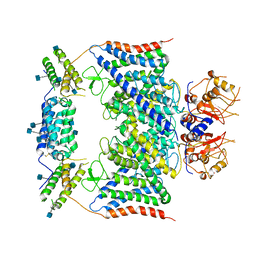 | | CLC-7/Ostm1 membrane protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H(+)/Cl(-) exchange transporter 7, Osteopetrosis-associated transmembrane protein 1 | | Authors: | Zhang, S.S, Yang, M.J. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insights into the human CLC-7/Ostm1 transporter.
Sci Adv, 6, 2020
|
|
7BB3
 
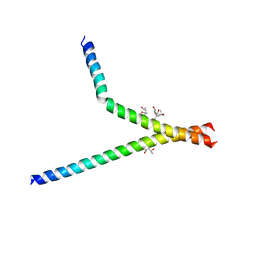 | | Structure of S. pombe YG-box oligomer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Survival motor neuron-like protein 1,Survival motor neuron-like protein 1 | | Authors: | Veepaschit, J, Grimm, C, Fischer, U. | | Deposit date: | 2020-12-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Identification and structural analysis of the Schizosaccharomyces pombe SMN complex.
Nucleic Acids Res., 49, 2021
|
|
4IFU
 
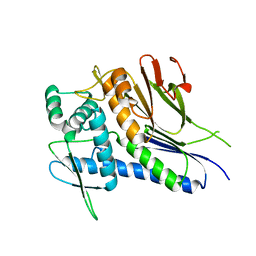 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, apo form | | Descriptor: | FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8334 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
6ZTL
 
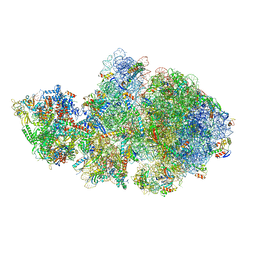 | | E. coli 70S-RNAP expressome complex in collided state bound to NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
4HB5
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR267. | | Descriptor: | Engineered Protein | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
