2R0N
 
 | | The effect of a Glu370Asp mutation in Glutaryl-CoA Dehydrogenase on Proton Transfer to the Dienolate Intermediate | | Descriptor: | 3-thiaglutaryl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Rao, K.S, Albro, M, Fu, Z, Narayanan, B, Baddam, S, Lee, H.J, Kim, J.J, Frerman, F.E. | | Deposit date: | 2007-08-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effect of a Glu370Asp mutation in glutaryl-CoA dehydrogenase on proton transfer to the dienolate intermediate.
Biochemistry, 46, 2007
|
|
2R0M
 
 | | The effect of a Glu370Asp Mutation in Glutaryl-CoA Dehydrogenase on Proton Transfer to the Dienolate Intermediate | | Descriptor: | 4-nitrobutanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Rao, K.S, Fu, Z, Albro, M, Narayanan, B, Baddam, S, Lee, H.J, Kim, J.J, Frerman, F.E. | | Deposit date: | 2007-08-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The effect of a Glu370Asp mutation in glutaryl-CoA dehydrogenase on proton transfer to the dienolate intermediate.
Biochemistry, 46, 2007
|
|
5I4N
 
 | |
2NO5
 
 | | Crystal Structure analysis of a Dehalogenase with intermediate complex | | Descriptor: | (2S)-2-CHLOROPROPANOIC ACID, (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, ... | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
3RTY
 
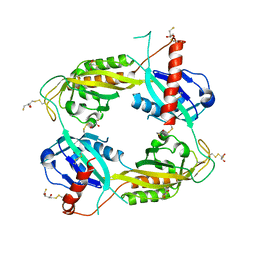 | | Structure of an Enclosed Dimer Formed by The Drosophila Period Protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Period circadian protein | | Authors: | King, H.A, Hoelz, A, Crane, B.R, Young, M.W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of an enclosed dimer formed by the Drosophila period protein.
J.Mol.Biol., 413, 2011
|
|
3D1K
 
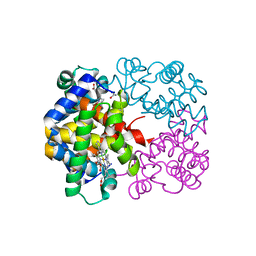 | | R/T intermediate quaternary structure of an antarctic fish hemoglobin in an alpha(CO)-beta(pentacoordinate) state | | Descriptor: | ACETYL GROUP, CARBON MONOXIDE, Hemoglobin subunit alpha-1, ... | | Authors: | Vitagliano, L, Vergara, A, Bonomi, G, Merlino, A, Mazzarella, L. | | Deposit date: | 2008-05-06 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Spectroscopic and crystallographic characterization of a tetrameric hemoglobin oxidation reveals structural features of the functional intermediate relaxed/tense state.
J.Am.Chem.Soc., 130, 2008
|
|
3O91
 
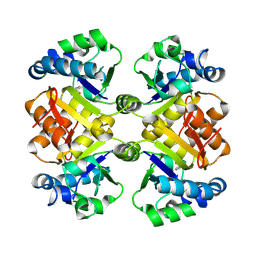 | | High resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into catalytic mechanism and inhibition by aldehydes | | Descriptor: | ZINC ION, nicotinamidase | | Authors: | French, J.B, Cen, Y, Sauve, A.A, Ealick, S.E. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | High-resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into the catalytic mechanism and inhibition by aldehydes .
Biochemistry, 49, 2010
|
|
3O90
 
 | | High resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into catalytic mechanism and inhibition by aldehydes | | Descriptor: | ZINC ION, nicotinamidase | | Authors: | French, J.B, Cen, Y, Sauve, A.A, Ealick, S.E. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | High-resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into the catalytic mechanism and inhibition by aldehydes .
Biochemistry, 49, 2010
|
|
2P04
 
 | |
3O92
 
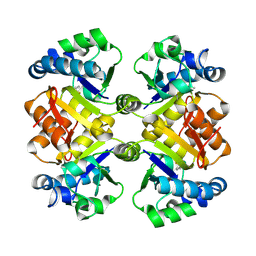 | | High resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into catalytic mechanism and inhibition by aldehydes | | Descriptor: | ZINC ION, nicotinamidase | | Authors: | French, J.B, Cen, Y, Sauve, A.A, Ealick, S.E. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into the catalytic mechanism and inhibition by aldehydes .
Biochemistry, 49, 2010
|
|
3O94
 
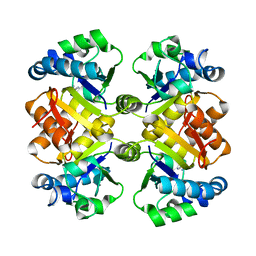 | | High resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into catalytic mechanism and inhibition by aldehydes | | Descriptor: | NICOTINAMIDE, ZINC ION, nicotinamidase | | Authors: | French, J.B, Cen, Y, Sauve, A.A, Ealick, S.E. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into the catalytic mechanism and inhibition by aldehydes .
Biochemistry, 49, 2010
|
|
2OWW
 
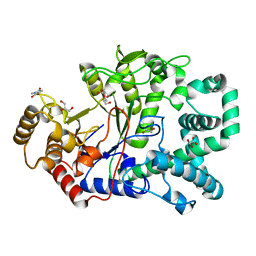 | | Covalent intermediate in amylomaltase in complex with the acceptor analog 4-deoxyglucose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-alpha-glucanotransferase, 4-deoxy-alpha-D-glucopyranose, ... | | Authors: | Barends, T.R.M, Bultema, J.B, Kaper, T, van der Maarel, M.J.E.C, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2007-02-17 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-way stabilization of the covalent intermediate in amylomaltase, an alpha-amylase-like transglycosylase.
J.Biol.Chem., 282, 2007
|
|
2OWC
 
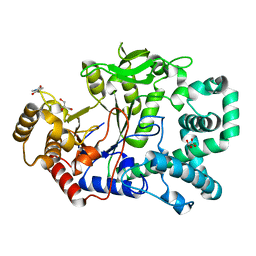 | | Structure of a covalent intermediate in Thermus thermophilus amylomaltase | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-alpha-glucanotransferase, GLYCEROL, ... | | Authors: | Barends, T.R.M, Bultema, J.B, Kaper, T, van der Maarel, M.J.E.C, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2007-02-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Three-way stabilization of the covalent intermediate in amylomaltase, an alpha-amylase-like transglycosylase.
J.Biol.Chem., 282, 2007
|
|
2PQ9
 
 | | E. coli EPSPS liganded with (R)-difluoromethyl tetrahedral reaction intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2ZCT
 
 | | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Hagihara, Y, Abe, M, Inoue, T, Yamamoto, T, Matsumura, H. | | Deposit date: | 2007-11-12 | | Release date: | 2008-05-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3O93
 
 | | High resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into catalytic mechanism and inhibition by aldehydes | | Descriptor: | ZINC ION, nicotinamidase | | Authors: | French, J.B, Cen, Y, Sauve, A.A, Ealick, S.E. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | High-resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into the catalytic mechanism and inhibition by aldehydes .
Biochemistry, 49, 2010
|
|
5TCU
 
 | | Methicillin sensitive Staphylococcus aureus 70S ribosome | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Eyal, Z, Ahmed, T, Belousoff, N, Mishra, S, Matzov, D, Bashan, A, Zimmerman, E, Lithgow, T, Bhushan, S, Yonath, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome.
MBio, 8, 2017
|
|
2P08
 
 | |
5RNT
 
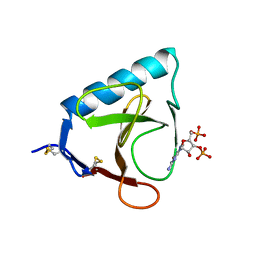 | | X-RAY ANALYSIS OF CUBIC CRYSTALS OF THE COMPLEX FORMED BETWEEN RIBONUCLEASE T1 AND GUANOSINE-3',5'-BISPHOSPHATE | | Descriptor: | GUANOSINE-3',5'-DIPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Saenger, W, Heinemann, U, Lenz, A. | | Deposit date: | 1991-03-28 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray analysis of cubic crystals of the complex formed between ribonuclease T1 and guanosine-3',5'-bisphosphate.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
2LBY
 
 | |
5DWX
 
 | |
2MY2
 
 | |
2DRW
 
 | | The crystal structutre of D-amino acid amidase from Ochrobactrum anthropi SV3 | | Descriptor: | BARIUM ION, D-Amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
2DNS
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with D-Phenylalanine | | Descriptor: | BARIUM ION, D-PHENYLALANINE, D-amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
1FP2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | 4'-HYDROXY-7-METHOXYISOFLAVONE, ISOFLAVONE O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-08-29 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat.Struct.Biol., 8, 2001
|
|
