8ILV
 
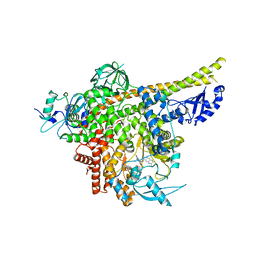 | | Cryo-EM structure of PI3Kalpha in complex with compound 18 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[3-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILU
 
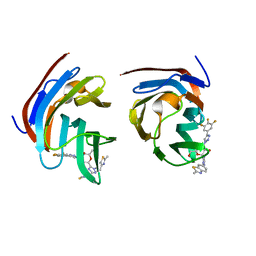 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | Authors: | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of benzothiazole derived monosaccharides as potent, selective, and orally bioavailable inhibitors of human and mouse galectin-3; a rare example of using a S···O binding interaction for drug design.
Bioorg.Med.Chem., 101, 2024
|
|
8ILS
 
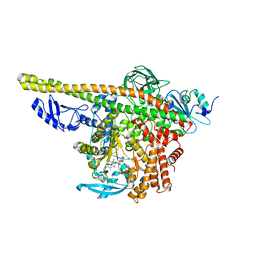 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILM
 
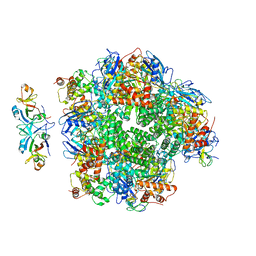 | | The cryo-EM structure of eight Rubisco large subunits (RbcL), two Arabidopsis thaliana Rubisco accumulation factors 1 (AtRaf1), and seven Arabidopsis thaliana Bundle Sheath Defective 2 (AtBSD2) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8ILL
 
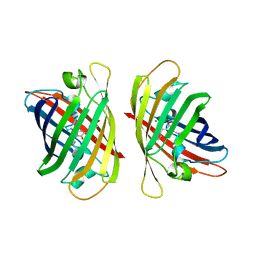 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | Descriptor: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
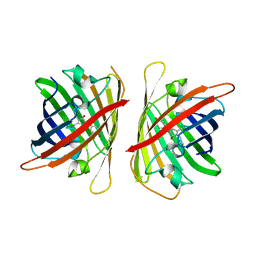 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | Descriptor: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILB
 
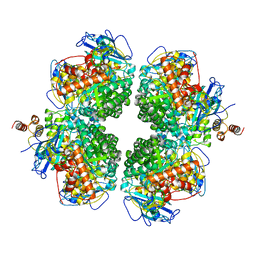 | | The complexes of RbcL, AtRaf1 and AtBSD2 (LFB) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | Descriptor: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | Authors: | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8IL8
 
 | |
8IL7
 
 | |
8IKZ
 
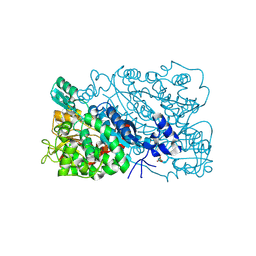 | | The mutant structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Bases of Dihydroxy Acid Dehydratase Inhibition and Biodesign for Self-Resistance.
Biodes Res, 6, 2024
|
|
8IKT
 
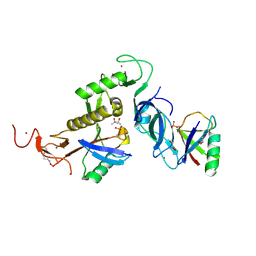 | | Ternary trans-complex of phospho-parkin with cis ACT and pUb | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-AMINOPROPANE, E3 ubiquitin-protein ligase parkin, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-03-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
8IKM
 
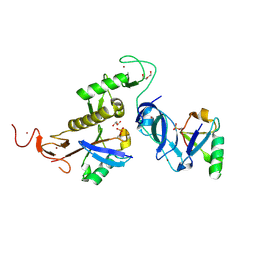 | | Trans complex of phospho parkin | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
8IKD
 
 | |
8IK8
 
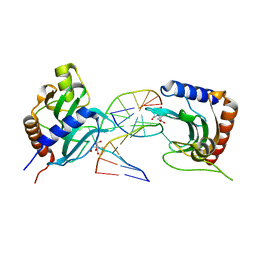 | |
8IK5
 
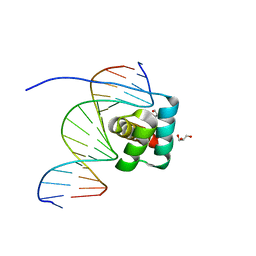 | | Transcription factor LMX1a homeobox domain in complex with Wnt1 promoter | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*AP*TP*TP*TP*AP*AP*TP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*AP*TP*TP*AP*AP*AP*TP*AP*TP*G)-3'), GLYCEROL, ... | | Authors: | Lv, M.Q, Lin, L.Q. | | Deposit date: | 2023-02-28 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Structural insights into the recognition of the A/T-rich motif in target gene promoters by the LMX1a homeobox domain
Febs J., 2024
|
|
8IK4
 
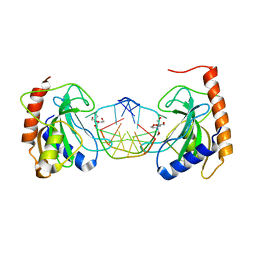 | |
8IK3
 
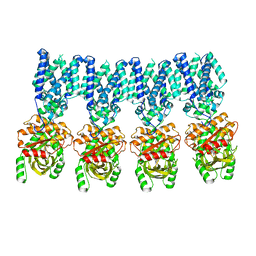 | |
8IK0
 
 | |
8IJY
 
 | | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, Deoxyribodipyrimidine photolyase-related protein, ... | | Authors: | Liu, Y, Xu, L, Zhang, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor
To Be Published
|
|
8IJX
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-18 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[(5-chloranyl-2-phenylmethoxy-phenyl)methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJW
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-06 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJV
 
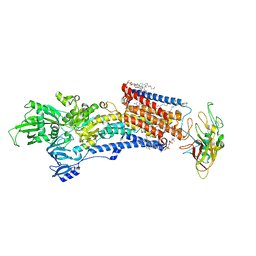 | | Cryo-EM structure of the gastric proton pump with bound DQ-02 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[[5-chloranyl-2-(4-chlorophenyl)phenyl]methoxy]-N-methyl-but-2-yn-1-amine, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJU
 
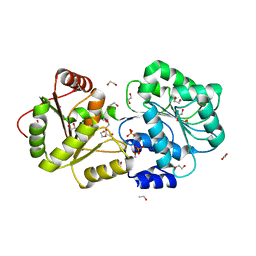 | | ATP-dependent RNA helicase DDX39A (URH49delta41) | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DDX39A, PHOSPHATE ION, ... | | Authors: | Mikami, B, Fujita, K, Masuda, S, Kojima, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural differences between the closely related RNA helicases, UAP56 and URH49, fashion distinct functional apo-complexes.
Nat Commun, 15, 2024
|
|
