9DLD
 
 | |
9DKH
 
 | |
9DLE
 
 | |
3EXU
 
 | | A glycoside hydrolase family 11 xylanase with an extended thumb region | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase, GLYCEROL | | Authors: | Vandermarliere, E, Pollet, A, Strelkov, S.V, Delcour, J.A, Courtin, C.M. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic and activity-based evidence for thumb flexibility and its relevance in glycoside hydrolase family 11 xylanases
Proteins, 77, 2009
|
|
9DKX
 
 | |
3FH5
 
 | |
9DMW
 
 | |
9DN7
 
 | |
9DJZ
 
 | |
9DKM
 
 | |
6UH1
 
 | | Structure of the EVA71 strain 11316 capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
3FBW
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant C176Y | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Dohnalek, J, Stsiapanava, A, Gavira, J.A, Kuta Smatanova, I, Kuty, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
9EJL
 
 | |
9EJK
 
 | |
9BI6
 
 | | Human proton sensing receptor GPR68 in complex with miniGsq | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Howard, M.K, Hoppe, N, Huang, X.P, Macdonald, C.B, Mehrotra, E, Rockefeller Grimes, P, Zahm, A.M, Trinidad, D.D, English, J, Coyote-Maestas, W, Manglik, A. | | Deposit date: | 2024-04-22 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of proton sensing by G protein-coupled receptors.
Cell, 188, 2025
|
|
3FH8
 
 | |
9BIP
 
 | | Human proton sensing receptor GPR4 in complex with miniGs | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Howard, M.K, Hoppe, N, Huang, X.P, Macdonald, C.B, Mehrotra, E, Rockefeller Grimes, P, Zahm, A.M, Trinidad, D.D, English, J, Coyote-Maestas, W, Manglik, A. | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular basis of proton sensing by G protein-coupled receptors.
Cell, 188, 2025
|
|
9BHM
 
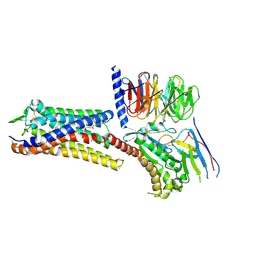 | | Human proton sensing receptor GPR68 in complex with miniGs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Howard, M.K, Hoppe, N, Huang, X.P, Macdonald, C.B, Mehrotra, E, Rockefeller Grimes, P, Zahm, A.M, Trinidad, D.D, English, J, Coyote-Maestas, W, Manglik, A. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of proton sensing by G protein-coupled receptors.
Cell, 188, 2025
|
|
1ZWB
 
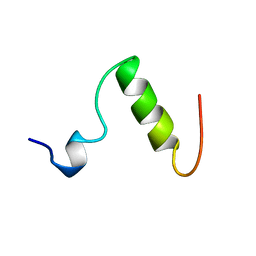 | |
1ZWE
 
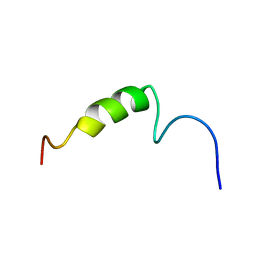 | |
3FAW
 
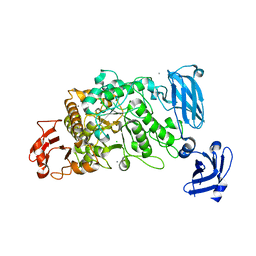 | | Crystal Structure of the Group B Streptococcus Pullulanase SAP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Reticulocyte binding protein | | Authors: | Gourlay, L.J. | | Deposit date: | 2008-11-18 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group B Streptococcus pullulanase crystal structures in the context of a novel strategy for vaccine development
J.Bacteriol., 191, 2009
|
|
1ZZZ
 
 | | Trypsin inhibitors with rigid tripeptidyl aldehydes | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3R)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, CALCIUM ION, TRYPSIN | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-06-02 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
2AAK
 
 | |
9BHL
 
 | | Human proton sensing receptor GPR65 in complex with miniGs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Nanobody 35, ... | | Authors: | Howard, M.K, Hoppe, N, Huang, X.P, Macdonald, C.B, Mehrotra, E, Rockefeller Grimes, P, Zahm, A.M, Trinidad, D.D, English, J, Coyote-Maestas, W, Manglik, A. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular basis of proton sensing by G protein-coupled receptors.
Cell, 188, 2025
|
|
3F5S
 
 | | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301
To be Published
|
|
