2JNC
 
 | | Refined 3D NMR structure of ECD1 of mCRF-R2beta at pH 5 | | Descriptor: | Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1QHZ
 
 | |
5OH1
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Aminoglutethimide | | Descriptor: | (3~{R})-3-(4-aminophenyl)-3-ethyl-piperidine-2,6-dione, Cereblon isoform 4, S-Thalidomide, ... | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5OH9
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thiazolidine-2,4-dione | | Descriptor: | 1,3-thiazole-2,4-dione, Cereblon isoform 4, ZINC ION | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
6ABQ
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
1PW9
 
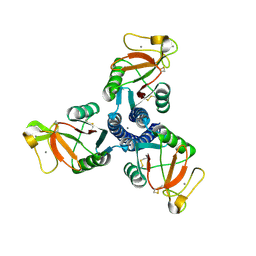 | | High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Shrive, A.K, Tharia, H.A, Strong, P, Kishore, U, Burns, I, Rizkallah, P.J, Reid, K.B, Greenhough, T.J. | | Deposit date: | 2003-07-01 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D
J.Mol.Biol., 331, 2003
|
|
2Y7L
 
 | |
4WO0
 
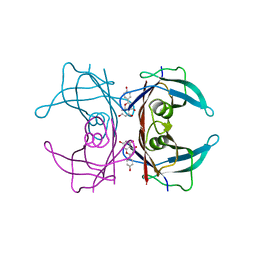 | | Crystal structure of transthyretin in complex with apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Folli, C, Berni, R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1LF7
 
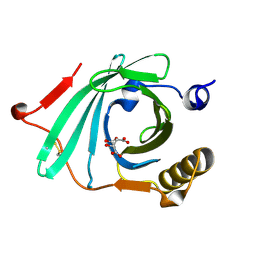 | | Crystal Structure of Human Complement Protein C8gamma at 1.2 A Resolution | | Descriptor: | CITRIC ACID, Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
6NFN
 
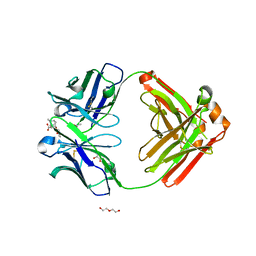 | | Fab fragment of anti-cocaine antibody h2E2 bound to benzoylecgonine | | Descriptor: | 3-(BENZOYLOXY)-8-METHYL-8-AZABICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pokkuluri, P.R, Tan, K. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7E2Y
 
 | | Serotonin-bound Serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E33
 
 | | Serotonin 1E (5-HT1E) receptor-Gi protein complex | | Descriptor: | 3-(1-methylpiperidin-4-yl)-1H-indol-5-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Z
 
 | | Aripiprazole-bound serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
4YH3
 
 | | Crystal structure of human BRD4(1) in complex with 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19a) | | Descriptor: | 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7E32
 
 | | Serotonin 1D (5-HT1D) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
4X5T
 
 | | alpha 1 glycine receptor transmembrane structure fused to the extracellular domain of GLIC | | Descriptor: | ACETATE ION, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Sauguet, L, Corringer, P.J, Huon, C, Delarue, M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Allosteric and hyperekplexic mutant phenotypes investigated on an alpha 1 glycine receptor transmembrane structure.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5ICR
 
 | | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine. | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Acyl-CoA synthase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Hung, D, Fisher, S.L, Edelstein, J, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-23 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine.
To Be Published
|
|
3FYO
 
 | | Crystal structure of the triple mutant (N23C/D247E/P249A) of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-manno-octulosonic acid 8-phosphate synthetase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Jameson, G.B, Parker, E.J, Cochrane, F.P, Patchett, M.L. | | Deposit date: | 2009-01-22 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversing evolution: re-establishing obligate metal ion dependence in a metal-independent KDO8P synthase
J.Mol.Biol., 390, 2009
|
|
2YLH
 
 | |
8E37
 
 | | Structure of Campylobacter concisus wild-type SeMet PglC | | Descriptor: | N,N'-diacetylbacilliosaminyl-1-phosphate transferase | | Authors: | Vuksanovic, N, Ray, L.C, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-08-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Synergistic computational and experimental studies of a phosphoglycosyl transferase membrane/ligand ensemble.
J.Biol.Chem., 299, 2023
|
|
1YZI
 
 | |
2Y7O
 
 | |
2Y7M
 
 | |
2Y7N
 
 | |
5NNJ
 
 | | Dimer structure of Sortilin ectodomain crystal form 3, 4.0 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Leloup, N.O.L, Janssen, B.J.C. | | Deposit date: | 2017-04-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
