6ON8
 
 | |
8E7F
 
 | | Crystal structure of the autotransporter Ssp from Serratia marcescens. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Extracellular serine protease, ... | | Authors: | Hor, L, Pilapitiya, A, Panjikar, S, Paxman, J.J, Heras, B. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a subtilisin-like autotransporter passenger domain reveals insights into its cytotoxic function.
Nat Commun, 14, 2023
|
|
6OKS
 
 | |
8E5N
 
 | | Structure of ARG1 complex with pyrrolidine-based non-boronic acid inhibitor 10 | | Descriptor: | 1-{[(3S,4S)-3-(3-fluorophenyl)-4-{[4-(1,3,4-triethyl-1H-pyrazol-5-yl)piperidin-1-yl]methyl}pyrrolidin-1-yl]methyl}cyclopentane-1-carboxylic acid, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L, Gathiaka, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Discovery of non-boronic acid Arginase 1 inhibitors through virtual screening and biophysical methods.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
5KCJ
 
 | | Structure of the human GluN1/GluN2A LBD in complex with GNE6901 | | Descriptor: | 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
5KGM
 
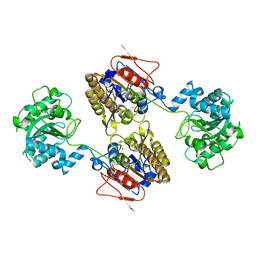 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
8E5M
 
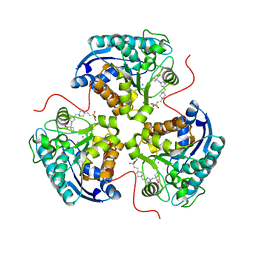 | | Structure of ARG1 complex with pyrrolidine-based non-boronic acid inhibitor 6 | | Descriptor: | 1-{[(3S,4S)-3-({4-[2-(4-fluorobenzene-1-sulfonyl)ethyl]piperidin-1-yl}methyl)-4-(3-fluorophenyl)pyrrolidin-1-yl]methyl}cyclopentane-1-carboxylic acid, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L, Gathiaka, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of non-boronic acid Arginase 1 inhibitors through virtual screening and biophysical methods.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
4WOE
 
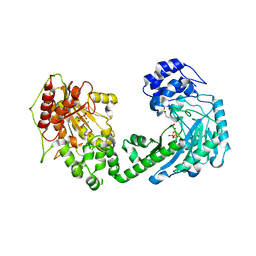 | | The duplicated taurocyamine kinase from Schistosoma mansoni with bound transition state analog (TSA) components | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
5KDM
 
 | |
8DMY
 
 | | Cryo-EM structure of cardiac muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8DMX
 
 | | Cryo-EM structure of skeletal muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8DNH
 
 | | Cryo-EM structure of nonmuscle beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8ED4
 
 | | Structure of the complex between the arsenite oxidase and its native electron acceptor cytochrome c552 from Pseudorhizobium sp. str. NT-26 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, AroA, AroB, ... | | Authors: | Maher, M.J, Poddar, N. | | Deposit date: | 2022-09-03 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of the complex between the arsenite oxidase from Pseudorhizobium banfieldiae sp. strain NT-26 and its native electron acceptor cytochrome c 552.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8EA8
 
 | | NKG2D complexed with inhibitor 4a | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5KGL
 
 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
8EA7
 
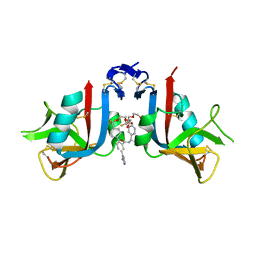 | | NKG2D complexed with inhibitor 3g | | Descriptor: | (4M)-N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4-(1-methyl-1H-pyrazol-5-yl)-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, DI(HYDROXYETHYL)ETHER, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA5
 
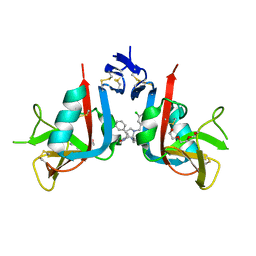 | | NKG2D complexed with inhibitor 1a | | Descriptor: | (3S,5aS,8aR)-3-benzyl-6-[(3,5-dichlorophenyl)methyl]-1,4-dimethyloctahydropyrrolo[3,2-e][1,4]diazepine-2,5-dione, NKG2-D type II integral membrane protein, TRIETHYLENE GLYCOL | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA9
 
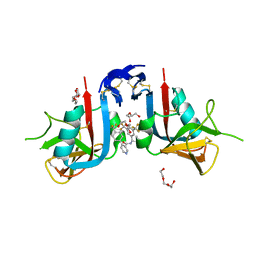 | | NKG2D complexed with inhibitor 4d | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-1-[3,5-bis(trifluoromethyl)phenyl]-2-(dimethylamino)-2-oxoethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAB
 
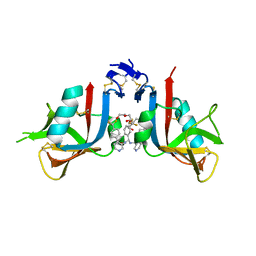 | | NKG2D complexed with inhibitor 4f | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(1S)-2-oxo-1-[3-(trifluoromethyl)phenyl]-2-({4-[4-(trifluoromethyl)phenyl]pyridin-3-yl}amino)ethyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EA6
 
 | | NKG2D complexed with inhibitor 3e | | Descriptor: | N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAA
 
 | | NKG2D complexed with inhibitor 4e | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-{(1S)-2-(dimethylamino)-1-[3-methyl-5-(trifluoromethyl)phenyl]-2-oxoethyl}-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4WL0
 
 | |
5KJ5
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169D in complex with NAD+ | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
6OI4
 
 | | RPN13 (19-132)-RPN2 (940-952) pY950-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, ... | | Authors: | Hemmis, C.W, Hill, C.P. | | Deposit date: | 2019-04-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13.
J.Biol.Chem., 294, 2019
|
|
5KLL
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived tautomeric intermediate in 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | (3~{E},5~{E})-6-oxidanyl-2-oxidanylidene-hexa-3,5-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
