1TOH
 
 | |
1R39
 
 | | THE STRUCTURE OF P38ALPHA | | Descriptor: | Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Patel, S.B, Cameron, P.M, Frantz-Wattley, B, O'Neill, E, Becker, J.W, Scapin, G. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lattice stabilization and enhanced diffraction in human p38 alpha crystals by protein engineering.
Biochim.Biophys.Acta, 1696, 2004
|
|
1R5Z
 
 | | Crystal Structure of Subunit C of V-ATPase | | Descriptor: | V-type ATP synthase subunit C | | Authors: | Iwata, M, Imamura, H, Stambouli, E, Ikeda, C, Tamakoshi, M, Nagata, K, Makyio, H, Hankamer, B, Barber, J, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1R9M
 
 | | Crystal Structure of Human Dipeptidyl Peptidase IV at 2.1 Ang. Resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, ... | | Authors: | Aertgeerts, K, Ye, S, Tennant, M.G, Collins, B, Rogers, J, Sang, B.C, Skene, R.J, Webb, D.R, Prasad, G.S. | | Deposit date: | 2003-10-30 | | Release date: | 2004-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation.
Protein Sci., 13, 2004
|
|
1T7L
 
 | |
1TVC
 
 | | FAD and NADH binding domain of methane monooxygenase reductase from Methylococcus capsulatus (Bath) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, METHANE MONOOXYGENASE COMPONENT C | | Authors: | Chatwood, L.L, Mueller, J, Gross, J.D, Wagner, G, Lippard, S.J. | | Deposit date: | 2004-06-29 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Flavin Domain from Soluble Methane Monooxygenase Reductase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
1TPL
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF TYROSINE PHENOL-LYASE | | Descriptor: | SULFATE ION, TYROSINE PHENOL-LYASE | | Authors: | Antson, A, Demidkina, T, Dauter, Z, Harutyunyan, E, Wilson, K. | | Deposit date: | 1992-11-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of tyrosine phenol-lyase.
Biochemistry, 32, 1993
|
|
1TKR
 
 | | Human Dipeptidyl Peptidase IV/CD26 inhibited with Diisopropyl FluoroPhosphate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tyrosine 547 Constitutes an Essential Part of the Catalytic Mechanism of Dipeptidyl Peptidase IV
J.Biol.Chem., 279, 2004
|
|
1SS7
 
 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-23 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
1TVL
 
 | |
1UTG
 
 | | REFINEMENT OF THE C2221 CRYSTAL FORM OF OXIDIZED UTEROGLOBIN AT 1.34 ANGSTROMS RESOLUTION | | Descriptor: | UTEROGLOBIN | | Authors: | Morize, I, Surcouf, E, Vaney, M.C, Buehner, M, Mornon, J.P. | | Deposit date: | 1989-04-03 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Refinement of the C222(1) crystal form of oxidized uteroglobin at 1.34 A resolution.
J.Mol.Biol., 194, 1987
|
|
1V71
 
 | | Crystal Structure of S.pombe Serine Racemase | | Descriptor: | Hypothetical protein C320.14 in chromosome III, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Goto, M. | | Deposit date: | 2003-12-09 | | Release date: | 2005-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of S.pombe Serine Racemase
to be published
|
|
1BVT
 
 | | METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS 569/H/9 | | Descriptor: | BICARBONATE ION, PROTEIN (BETA-LACTAMASE), ZINC ION | | Authors: | Carfi, A, Duee, E, Dideberg, O. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 A resolution structure of the zinc (II) beta-lactamase from Bacillus cereus.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
5CJF
 
 | | The crystal structure of the human carbonic anhydrase XIV in complex with a 1,1'-biphenyl-4-sulfonamide inhibitor. | | Descriptor: | 4'-(4-aminobenzoyl)biphenyl-4-sulfonamide, Carbonic anhydrase 14, GLYCEROL, ... | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2015-07-14 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of 1,1'-Biphenyl-4-sulfonamides as a New Class of Potent and Selective Carbonic Anhydrase XIV Inhibitors.
J.Med.Chem., 58, 2015
|
|
1CQZ
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE. | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5GJA
 
 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with 2-PA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRIDINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
5JY0
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with substrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Asp/Glu-specific dipeptidyl-peptidase, LEU-ASP-VAL | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
1CKV
 
 | |
1COW
 
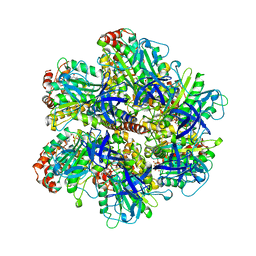 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
5JWF
 
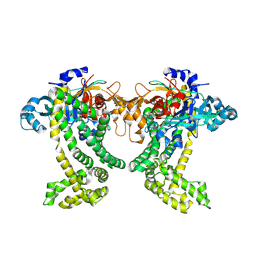 | | Crystal structure of Porphyromonas gingivalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
6WTI
 
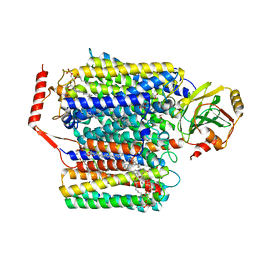 | | The Cryo-EM structure of the ubiquinol oxidase from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome o ubiquinol oxidase, ... | | Authors: | Su, C.-C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
5K7K
 
 | |
1D5V
 
 | | SOLUTION STRUCTURE OF THE FORKHEAD DOMAIN OF THE ADIPOCYTE-TRANSCRIPTION FACTOR FREAC-11 (S12) | | Descriptor: | S12 TRANSCRIPTION FACTOR (FKH-14) | | Authors: | van Dongen, M.J.P, Cederberg, A, Carlsson, P, Enerback, S, Wikstrom, M. | | Deposit date: | 1999-10-12 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the DNA-binding domain of the adipocyte-transcription factor FREAC-11.
J.Mol.Biol., 296, 2000
|
|
1DB6
 
 | | SOLUTION STRUCTURE OF THE DNA APTAMER 5'-CGACCAACGTGTCGCCTGGTCG-3' COMPLEXED WITH ARGININAMIDE | | Descriptor: | ARGININEAMIDE, DNA | | Authors: | Robertson, S.A, Harada, K, Frankel, A.D, Wemmer, D.E. | | Deposit date: | 1999-11-02 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination and binding kinetics of a DNA aptamer-argininamide complex.
Biochemistry, 39, 2000
|
|
5JXP
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
