2XP8
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-(MORPHOLIN-4-YLCARBONYL)-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XSD
 
 | | Crystal Structure of the dimeric Oct-6 (Pou3f1) POU domain bound to palindromic MORE DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*TP*GP*CP*AP*TP*GP*AP*GP*GP*AP)-3', 5'-D(*CP*CP*TP*CP*AP*TP*GP*CP*AP*TP*AP)-3', ... | | Authors: | Jauch, R, Choo, S.H, Ng, C.K.L, Kolatkar, P.R. | | Deposit date: | 2010-09-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Crystal Structure of the Dimeric Oct6 (POU3F1) POU Domain Bound to Palindromic More DNA.
Proteins, 79, 2011
|
|
2XG8
 
 | |
2XNB
 
 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 3,4-DIMETHYL-5-(2-{[(1Z)-4-PIPERAZIN-1-YLCYCLOHEXA-2,4-DIEN-1-YLIDENE]AMINO}PYRIMIDIN-4-YL)-1,3-THIAZOL-2(3H)-ONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-08-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
2XKP
 
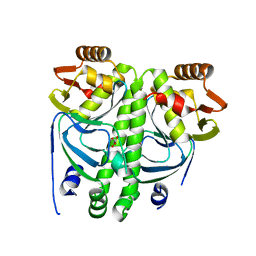 | |
6I7M
 
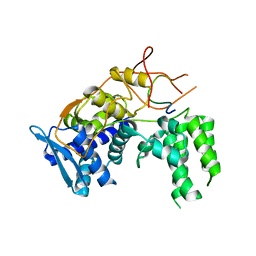 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 4. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
2XMY
 
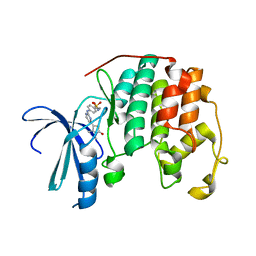 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 4-[4-(3,4-DIMETHYL-2-OXO-2,3-DIHYDRO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-N-(2-METHOXY-ETHYL)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
2XP9
 
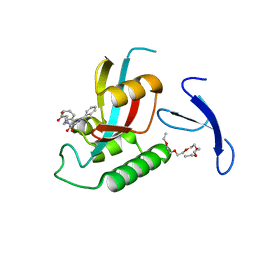 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-[BENZYL(CARBOXYMETHYL)CARBAMOYL]-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XP5
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 5-METHYL-2-PHENYL-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XP3
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 5-(2-METHOXYPHENYL)-2-FUROIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XPA
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-[(2-amino-2-oxoethyl)(methyl)carbamoyl]-2-phenyl-1H-imidazole-5-carboxylic acid, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XJY
 
 | | Crystal structure of the LMO2:LDB1-LID complex, P21 crystal form | | Descriptor: | LIM DOMAIN-BINDING PROTEIN 1, RHOMBOTIN-2, ZINC ION | | Authors: | El Omari, K, Karia, D, Porcher, C, Mancini, E.J. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Leukemia Oncogene Lmo2: Implications for the Assembly of a Hematopoietic Transcription Factor Complex.
Blood, 117, 2011
|
|
5LUQ
 
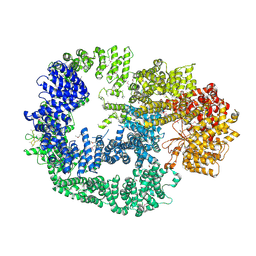 | | Crystal Structure of Human DNA-dependent Protein Kinase Catalytic Subunit (DNA-PKcs) | | Descriptor: | C-terminal fragment of KU80 (KU80ct194), DNA-dependent protein kinase catalytic subunit,DNA-dependent Protein Kinase Catalytic Subunit,DNA-dependent protein kinase catalytic subunit | | Authors: | Sibanda, B.L, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-15 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair.
Science, 355, 2017
|
|
2XP4
 
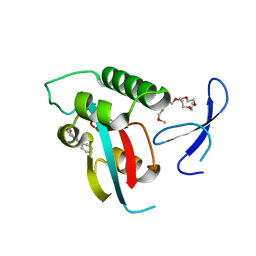 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 2-phenyl-1H-imidazole-4-carboxylic acid, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XJZ
 
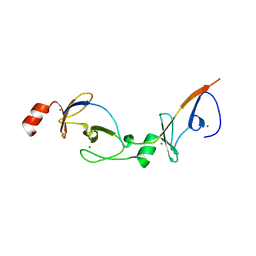 | | Crystal structure of the LMO2:LDB1-LID complex, C2 crystal form | | Descriptor: | CHLORIDE ION, LIM DOMAIN-BINDING PROTEIN 1, RHOMBOTIN-2, ... | | Authors: | El Omari, K, Karia, D, Porcher, C, Mancini, E.J. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Leukemia Oncogene Lmo2: Implications for the Assembly of a Hematopoietic Transcription Factor Complex.
Blood, 117, 2011
|
|
4F7M
 
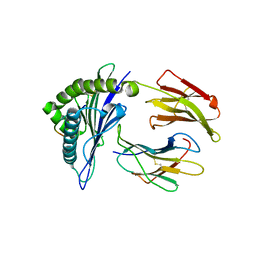 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PA (649-658) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, George, F.G. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
7SUD
 
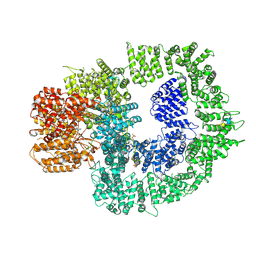 | | CryoEM structure of DNA-PK complex VIII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SU3
 
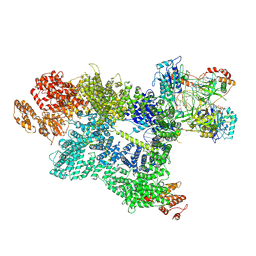 | | CryoEM structure of DNA-PK complex VII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
6H9G
 
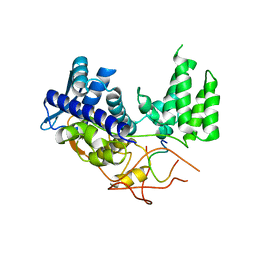 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 1. | | Descriptor: | Nucleoprotein, Polypeptide loop | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Munier, S, Carlero, D, Naffakh, N, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-08-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
8HXZ
 
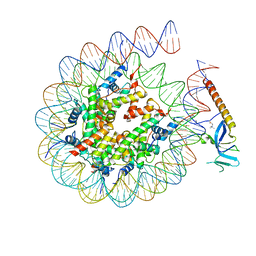 | | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (352-MER), Histone H2A, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6I7B
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
7UPN
 
 | |
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
4XMN
 
 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XMM
 
 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
