5FUJ
 
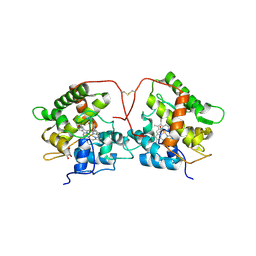 | |
7S8Q
 
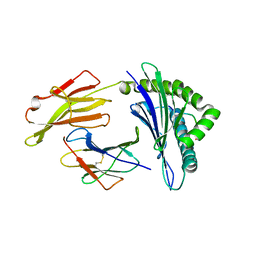 | | Crystal Structure of HLA A*1101 in complex with KTNGNAFIGK, an 10-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
7SIU
 
 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
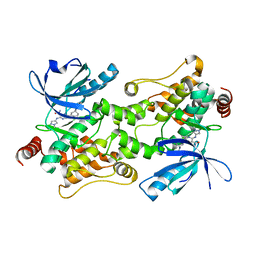
7SJY
 
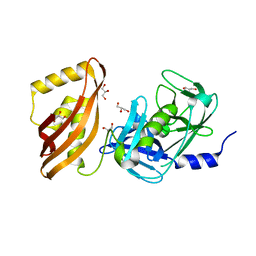 | |
7SKR
 
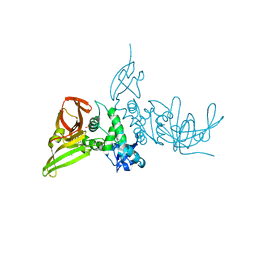 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor 37 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(2-methoxypyridin-4-yl)methyl]-2-[(1R)-1-(naphthalen-1-yl)ethyl]-2-azaspiro[3.3]heptane-6-carboxamide, ... | | Authors: | Durie, I, Shepard, J, Freitas, B, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
7SSU
 
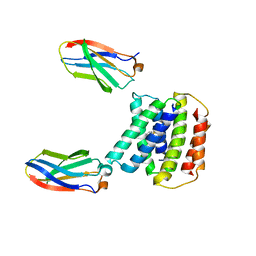 | |
5FBK
 
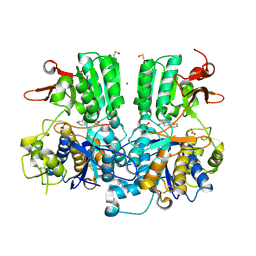 | | Crystal structure of the extracellular domain of human calcium sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
7SNZ
 
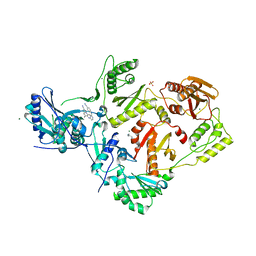 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 6-((2-((4-cyanophenyl)amino)pyrimidin-4-yl)amino)-5,7-dimethylindolizine-2-carbonitrile (JLJ604) | | Descriptor: | (4R)-6-{[2-(4-cyanoanilino)pyrimidin-4-yl]amino}-5,7-dimethylindolizine-2-carbonitrile, 1,4-DIAMINOBUTANE, MAGNESIUM ION, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7S5F
 
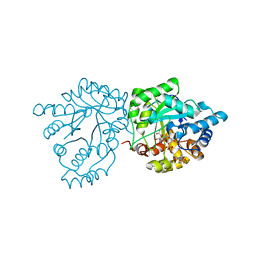 | | Crystal structure of mannose-6-phosphate reductase from celery (Apium graveolens) leaves with NADP+ and mannonic acid bound | | Descriptor: | D-MANNONIC ACID, Manose-6-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
7SNP
 
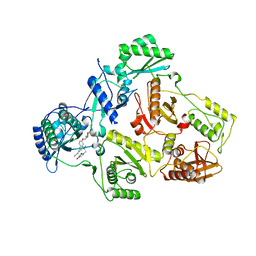 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-(2-morpholinoethoxy)phenoxy)phenyl)acrylonitrile (JLJ530) | | Descriptor: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-[2-(morpholin-4-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO2
 
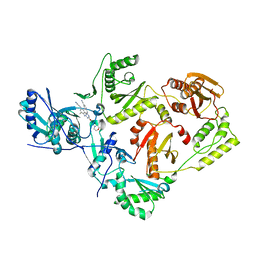 | | Crystal Structure of HIV-1 Reverse Transcriptase Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SMJ
 
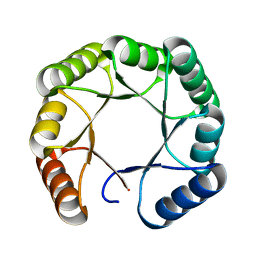 | |
7SO1
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.727 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO3
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase K103N/Y181C Variant in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.767 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SGO
 
 | |
7S5I
 
 | | Crystal structure of Aldose-6-phosphate reductase (Ald6PRase) from peach (Prunus persica) leaves | | Descriptor: | Sorbitol-6-phosphate dehydrogenase | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
7SGN
 
 | |
7SGP
 
 | |
7SFA
 
 | |
7SF9
 
 | |
1ZVW
 
 | | The Crystal Structure of TrpD (Rv2192c) from Mycobacterium tuberculosis in Complex with PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, BENZAMIDINE, ... | | Authors: | Lee, C.E, Lott, J.S, Baker, E.N, Arcus, V.L, Javid-Majd, F, Goodfellow, C, Hung, L.-W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-06-02 | | Release date: | 2006-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of TrpD, a Metabolic Enzyme Essential for Lung Colonization by Mycobacterium tuberculosis, in Complex with its Substrate Phosphoribosylpyrophosphate.
J.Mol.Biol., 355, 2006
|
|
7S5O
 
 | |
7SH6
 
 | |
7RZB
 
 | | BrxA from Staphylococcus aureus with bacillithiol mixed disulfide | | Descriptor: | (2S)-2-{[2-(L-cysteinylamino)-2-deoxy-alpha-D-glucopyranosyl]oxy}butanedioic acid, Bacillithiol system oxidoreductase, YphP/YqiW family, ... | | Authors: | Cook, P.D, McHugh, C.S. | | Deposit date: | 2021-08-27 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of BrxA from Staphylococcus aureus, a bacilliredoxin involved in redox homeostasis in Firmicutes.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7SMU
 
 | | Crystal Structure of Consomatin-Ro1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45-pentadecaoxaoctatetracontane-1,48-diol, Consomatin-Ro1 | | Authors: | Ramiro, I.B.L, Whitby, F.G, Hill, C.P, Safavi-Hemami, H, Concepcion, G.P, Olivera, B.M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads.
Sci Adv, 8, 2022
|
|
