1RE2
 
 | | HUMAN LYSOZYME LABELLED WITH TWO 2',3'-EPOXYPROPYL BETA-GLYCOSIDE OF N-ACETYLLACTOSAMINE | | Descriptor: | GLYCEROL, PROTEIN (LYSOZYME), beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1998-11-05 | | Release date: | 1999-05-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual affinity labeling of the active site of human lysozyme with an N-acetyllactosamine derivative: first ligand assisted recognition of the second ligand.
Biochemistry, 38, 1999
|
|
1MUM
 
 | |
3GOM
 
 | | Barium bound to the Holliday junction sequence d(TCGGCGCCGA)4 | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3', BARIUM ION | | Authors: | Naseer, A, Cardin, C.J. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5IRZ
 
 | | Structure of TRPV1 determined in lipid nanodisc | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4S,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(pentanoyloxy)propan-2-yl decanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, (4R,7S)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(pentanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphatetradecan-1-aminium, ... | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
1QJB
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 1) | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
3HHN
 
 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | Descriptor: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | Deposit date: | 2009-05-15 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.987 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
1TDU
 
 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE (DURD) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE, PHOSPHATE ION, ... | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-25 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1TJS
 
 | | E. COLI THYMIDYLATE SYNTHASE | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-27 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1TRG
 
 | | E. COLI THYMIDYLATE SYNTHASE IN SYMMETRIC COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1998-05-21 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
2OIU
 
 | | L1 Ribozyme Ligase circular adduct | | Descriptor: | L1 Ribozyme RNA Ligase, MAGNESIUM ION | | Authors: | Robertson, M.P, Scott, W.G. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of ribozyme-catalyzed RNA assembly.
Science, 315, 2007
|
|
1LSU
 
 | | KTN Bsu222 Crystal Structure in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Conserved hypothetical protein yuaA | | Authors: | Roosild, T.P, Miller, S, Booth, I.R, Choe, S. | | Deposit date: | 2002-05-18 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch.
Cell(Cambridge,Mass.), 109, 2002
|
|
7SA8
 
 | |
6EA5
 
 | | Structure of BDBV GPcl in complex with the pan-ebolavirus mAb ADI-15878 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-15878 Fab Heavy Chain, ... | | Authors: | King, L.B, West, B.R, Moyer, C.L, Fusco, M.L, Milligan, J.C, Hui, S, Saphire, E.O. | | Deposit date: | 2018-08-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.75 Å) | | Cite: | Structural Basis of Pan-Ebolavirus Neutralization by a Human Antibody against a Conserved, yet Cryptic Epitope.
MBio, 9, 2018
|
|
1M7O
 
 | | Plasmodium Falciparum Triosephosphate isomerase (PfTIM) compled to substrate analog 3-phosphoglycerate (3PG) | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Triosephosphate Isomerase | | Authors: | Parthasarathy, S, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Plasmodium falciparum triosephosphate isomerase complexed to substrate analogues: observation of the catalytic loop in the open conformation in the ligand-bound state.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HCL
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
1DB1
 
 | | CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D NUCLEAR RECEPTOR | | Authors: | Rochel, N, Wurtz, J.M, Mitschler, A, Klaholz, B, Moras, D. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand.
Mol.Cell, 5, 2000
|
|
6EWM
 
 | | Crystal structure of heme free PORPHYROMONAS GINGIVALIS HEME-BINDING PROTEIN HMUY | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Haemophore HmuY, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Bielecki, M, Olczak, T, Olczak, M. | | Deposit date: | 2017-11-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tannerella forsythiaTfo belongs toPorphyromonas gingivalisHmuY-like family of proteins but differs in heme-binding properties.
Biosci. Rep., 38, 2018
|
|
1HCK
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HUMAN CYCLIN-DEPENDENT KINASE 2, MAGNESIUM ION | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
1ML7
 
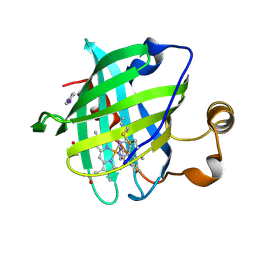 | | Crystal structure of nitrophorin 4 complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, 5,8-DIMETHYL-1,2,3,4-TETRAVINYLPORPHINE-6,7-DIPROPIONIC ACID FERROUS COMPLEX, nitrophorin 4 | | Authors: | Berry, R.E, Ding, X.D, Weichsel, A, Montfort, W.R, Walker, F.A. | | Deposit date: | 2002-08-30 | | Release date: | 2002-09-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Axial ligand complexes of the Rhodnius nitrophorins: reduction potentials, binding constants, EPR spectra, and structures of the 4-iodopyrazole and imidazole complexes of NP4
J.Biol.Inorg.Chem., 9, 2004
|
|
6F1S
 
 | | C-terminal domain of CglI restriction endonuclease H subunit | | Descriptor: | 1,2-ETHANEDIOL, CglIIR protein, FORMIC ACID | | Authors: | Tamulaitiene, G, Grigaitis, R, Zaremba, M, Silanskas, A. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-14 | | Last modified: | 2019-01-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The H-subunit of the restriction endonuclease CglI contains a prototype DEAD-Z1 helicase-like motor.
Nucleic Acids Res., 46, 2018
|
|
4A11
 
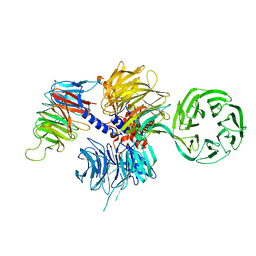 | | Structure of the hsDDB1-hsCSA complex | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, DNA EXCISION REPAIR PROTEIN ERCC-8 | | Authors: | Bohm, K, Scrima, A, Fischer, E.S, Gut, H, Thomae, N.H. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
6V4B
 
 | | DeCLIC N-terminal Domain 34-202 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Neur_chan_LBD domain-containing protein, ... | | Authors: | Delarue, M, Hu, H.D. | | Deposit date: | 2019-11-27 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for allosteric transitions of a multidomain pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4A08
 
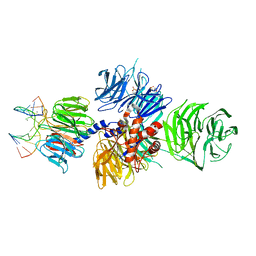 | | Structure of hsDDB1-drDDB2 bound to a 13 bp CPD-duplex (purine at D-1 position) at 3.0 A resolution (CPD 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*CP*GP*CP*GP*AP*(TTD)P*GP*CP*GP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*CP*GP*CP*CP*CP*TP*CP*GP*CP*G)-3', ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0A
 
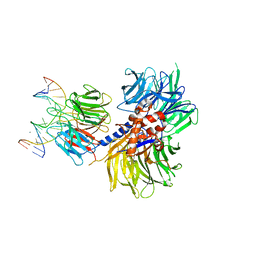 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A09
 
 | | Structure of hsDDB1-drDDB2 bound to a 15 bp CPD-duplex (purine at D-1 position) at 3.1 A resolution (CPD 2) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*GP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
