5D2M
 
 | | Complex between human SUMO2-RANGAP1, UBC9 and ZNF451 | | Descriptor: | 1,2-ETHANEDIOL, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Cappadocia, L, Lima, C.D. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7R5W
 
 | | Crystal structure of YTHDF2 with compound YLI_DF_029 | | Descriptor: | 6-cyclopropyl-1H-pyrimidine-2,4-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
4TZC
 
 | | Crystal Structure of Murine Cereblon in Complex with Thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
2ZDL
 
 | | Exploring trypsin S3 pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentyloxy)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Baum, B, Brandt, T, Heine, A, Klebe, G. | | Deposit date: | 2007-11-26 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
2ZQ2
 
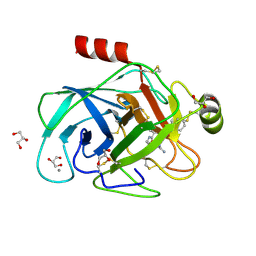 | | Exploring trypsin S3 pocket | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-08-03 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
1A6N
 
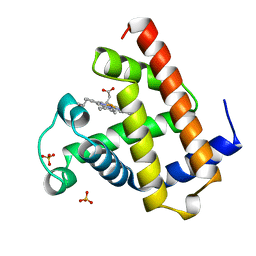 | | DEOXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
6OQQ
 
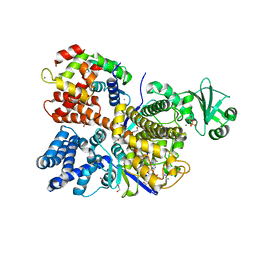 | | Legionella pneumophila SidJ/Saccharomyces cerevisiae calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Black, M, Osinski, A. | | Deposit date: | 2019-04-28 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases.
Science, 364, 2019
|
|
4GPO
 
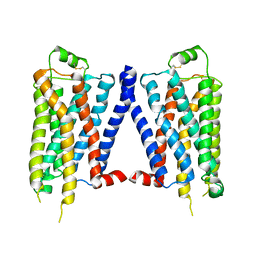 | |
2OVD
 
 | | Crystal Structure of Human Complement Protein C8gamma with Laurate | | Descriptor: | Complement component 8, gamma polypeptide, LAURIC ACID | | Authors: | Chiswell, B, Lovelace, L.L, Brannen, C, Ortlund, E.A, Lebioda, L, Sodetz, J.M. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural features of the ligand binding site on human complement protein C8gamma: A member of the lipocalin family
Biochim.Biophys.Acta, 1774, 2007
|
|
6TRX
 
 | | Crystal structure of DPP8 in complex with 1G244 | | Descriptor: | (2~{S})-2-azanyl-4-[4-[bis(4-fluorophenyl)methyl]piperazin-1-yl]-1-(1,3-dihydroisoindol-2-yl)butane-1,4-dione, Dipeptidyl peptidase 8, PHOSPHATE ION, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2019-12-19 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Aerosol-based ligand soaking of reservoir-free protein crystals.
J.Appl.Crystallogr., 54, 2021
|
|
1SL5
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SL4
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
2GI4
 
 | |
6MU2
 
 | | Structure of full-length IP3R1 channel in the Apo-state | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
1PW9
 
 | | High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Shrive, A.K, Tharia, H.A, Strong, P, Kishore, U, Burns, I, Rizkallah, P.J, Reid, K.B, Greenhough, T.J. | | Deposit date: | 2003-07-01 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D
J.Mol.Biol., 331, 2003
|
|
6A06
 
 | | Structure of pSTING complex | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
 | | Structure of pSTING complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
1GMM
 
 | | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | | Descriptor: | CALCIUM ION, CBM6, SODIUM ION, ... | | Authors: | Czjzek, M, Mosbah, A, Bolam, D, Allouch, J, Zamboni, V, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2001-09-19 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved.
J.Biol.Chem., 276, 2001
|
|
3VHB
 
 | | IMIDAZOLE ADDUCT OF THE BACTERIAL HEMOGLOBIN FROM VITREOSCILLA SP. | | Descriptor: | IMIDAZOLE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Boffi, A, Coletta, M, Mozzarelli, A, Pesce, A, Tarricone, C, Ascenzi, P. | | Deposit date: | 1999-03-17 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anticooperative ligand binding properties of recombinant ferric Vitreoscilla homodimeric hemoglobin: a thermodynamic, kinetic and X-ray crystallographic study.
J.Mol.Biol., 291, 1999
|
|
6JLN
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
3DCG
 
 | | Crystal Structure of the HIV Vif BC-box in Complex with Human ElonginB and ElonginC | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Stanley, B.J, Ehrlich, E.S, Short, L, Yu, Y, Xiao, Z, Yu, X.-F, Xiong, Y. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly
J.Virol., 82, 2008
|
|
1LF7
 
 | | Crystal Structure of Human Complement Protein C8gamma at 1.2 A Resolution | | Descriptor: | CITRIC ACID, Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
1QHZ
 
 | |
1SL6
 
 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1DQG
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE RICH DOMAIN OF MANNOSE RECEPTOR | | Descriptor: | MANNOSE RECEPTOR, SULFATE ION | | Authors: | Liu, Y, Chirino, A.J, Misulovin, Z, Leteux, C, Feizi, T, Nussenzweig, M.C, Bjorkman, P.J. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cysteine-rich domain of mannose receptor complexed with a sulfated carbohydrate ligand.
J.Exp.Med., 191, 2000
|
|
