5VLG
 
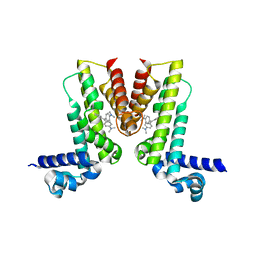 | | Crystal structure of EilR in complex with malachite green | | Descriptor: | MALACHITE GREEN, Regulatory protein TetR | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.932 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
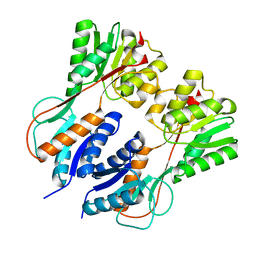
3HS3
 
 | |
2O20
 
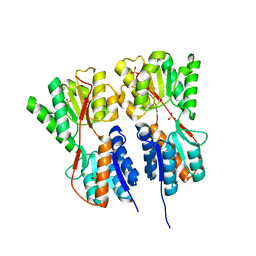 | | Crystal structure of transcription regulator CcpA of Lactococcus lactis | | Descriptor: | CHLORIDE ION, Catabolite control protein A, SULFATE ION | | Authors: | Loll, B, Kowalczyk, M, Alings, C, Chieduch, A, Bardowski, J, Saenger, W, Biesiadka, J. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the transcription regulator CcpA from Lactococcus lactis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
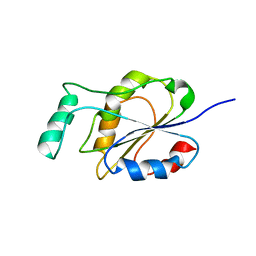
2MWM
 
 | |
2N1M
 
 | |
2WQF
 
 | | Crystal Structure of the Nitroreductase CinD from Lactococcus lactis in Complex with FMN | | Descriptor: | COPPER INDUCED NITROREDUCTASE D, FLAVIN MONONUCLEOTIDE | | Authors: | Waltersperger, S.M, Oberholzer, A.E, Solioz, M, Baumann, U. | | Deposit date: | 2009-08-20 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Function of Cind (Ytjd) of Lactococcus Lactis, a Copper-Induced Nitroreductase Involved in Defense Against Oxidative Stress.
J.Bacteriol., 192, 2010
|
|
3EDO
 
 | |
2IU6
 
 | | REGULATION OF THE DHA OPERON OF LACTOCOCCUS LACTIS | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCEROL | | Authors: | Srinivas, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2006-05-27 | | Release date: | 2006-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regulation of the Dha Operon of Lactococcus Lactis: A Deviation from the Rule Followed by the Tetr Family of Transcription Regulators
J.Biol.Chem., 281, 2006
|
|
1UXC
 
 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1UXD
 
 | | Fructose repressor DNA-binding domain, NMR, 34 structures | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
7PLE
 
 | | ArsH of Paracoccus denitrificans | | Descriptor: | NADPH-dependent FMN reductase | | Authors: | Kryl, M. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The ArsH Protein Product of the Paracoccus denitrificans ars Operon Has an Activity of Organoarsenic Reductase and Is Regulated by a Redox-Responsive Repressor.
Antioxidants, 11, 2022
|
|
3KEO
 
 | | Crystal Structure of a Rex-family transcriptional regulatory protein from Streptococcus agalactiae complexed with NAD+ | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thiyagarajan, S, Logan, D, von Wachenfeldt, C. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence.
Plos Pathog., 17, 2021
|
|
3KET
 
 | | Crystal structure of a Rex-family transcriptional regulatory protein from Streptococcus agalactiae bound to a palindromic operator | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*TP*CP*AP*CP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Thiyagarajan, S, Logan, D, von Wachenfeldt, C. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence.
Plos Pathog., 17, 2021
|
|
3KEQ
 
 | |
3V2U
 
 | | Crystal structure of the yeast GAL regulon complex of the repressor, Gal80p, and the transducer, Gal3p, with galactose and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Galactose/lactose metabolism regulatory protein GAL80, ... | | Authors: | Lavy, T, Kumar, P.R, He, H, Joshua-Tor, L. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.
Genes Dev., 26, 2012
|
|
1EA4
 
 | | TRANSCRIPTIONAL REPRESSOR COPG/22bp dsDNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*GP*TP*GP *CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*TP*C)-3'), DNA(5'-D(*AP*GP*AP*TP*TP*GP*CP*AP*TP *TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*GP*TP*T)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Costa, M, Sola, M, Acebo, P, Eritja, R, Espinosa, M, Solar, G.D, Coll, M. | | Deposit date: | 2000-11-05 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasmid Transcriptional Repressor Copg Oligomerises to Render Helical Superstructures Unbound and in Complexes with Oligonucleotides
J.Mol.Biol., 310, 2001
|
|
8EXP
 
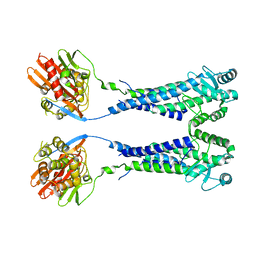 | | Cryo-EM structure of S. aureus BlaR1 with C2 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXS
 
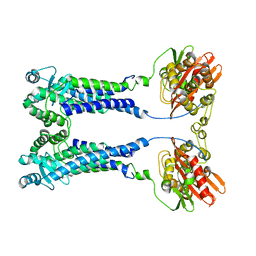 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXT
 
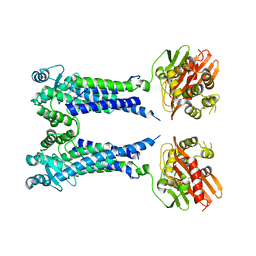 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant in complex with ampicillin | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXQ
 
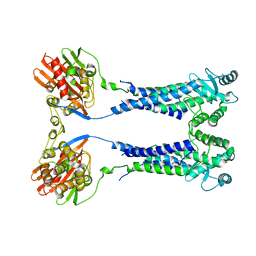 | | Cryo-EM structure of S. aureus BlaR1 with C1 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
1XSD
 
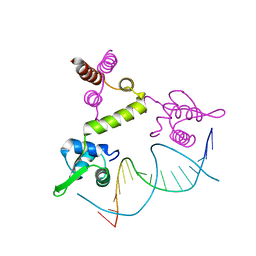 | | Crystal structure of the BlaI repressor in complex with DNA | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', penicillinase repressor | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Robinson, H, Scarsdale, N, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2004-10-19 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1B28
 
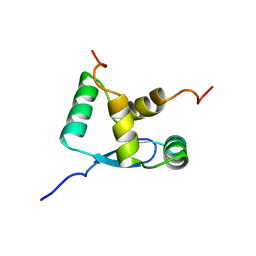 | | ARC REPRESSOR MYL MUTANT FROM SALMONELLA BACTERIOPHAGE P22 | | Descriptor: | PROTEIN (REGULATORY PROTEIN ARC) | | Authors: | Rietveld, A.W.M, Nooren, I.M.A, Sauer, R.T, Kaptein, R, Boelens, R. | | Deposit date: | 1998-12-05 | | Release date: | 1999-11-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of an Arc repressor mutant reveal premelting conformational changes related to DNA binding.
Biochemistry, 38, 1999
|
|
1B01
 
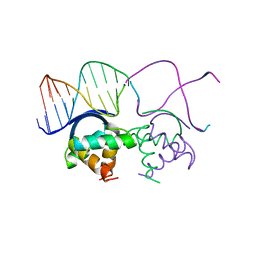 | | TRANSCRIPTIONAL REPRESSOR COPG/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*GP*CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*GP*CP*AP*TP*TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*G)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Sola, M, Acebo, P, Parraga, A, Guasch, A, Eritja, R, Gonzalez, A, Espinosa, M, del Solar, G, Coll, M. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator.
EMBO J., 17, 1998
|
|
2CPG
 
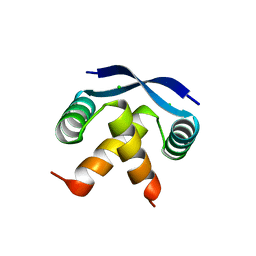 | | TRANSCRIPTIONAL REPRESSOR COPG | | Descriptor: | CHLORIDE ION, TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Sola, M, Acebo, P, Parraga, A, Guasch, A, Eritja, R, Gonzalez, A, Espinosa, M, del Solar, G, Coll, M. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator.
EMBO J., 17, 1998
|
|
2IU5
 
 | | Dihydroxyacetone kinase operon activator DhaS | | Descriptor: | HTH-TYPE DHAKLM OPERON TRANSCRIPTIONAL ACTIVATOR DHAS | | Authors: | Srinivas, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2006-05-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulation of the Dha Operon of Lactococcus Lactis: A Deviation from the Rule Followed by the Tetr Family of Transcription Regulators
J.Biol.Chem., 281, 2006
|
|
