4ZRJ
 
 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4YL8
 
 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
2D10
 
 | |
2D11
 
 | |
1NI2
 
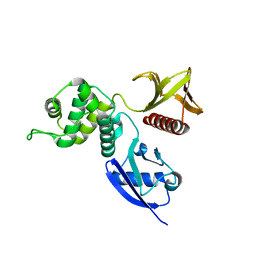 | | Structure of the active FERM domain of Ezrin | | Descriptor: | Ezrin | | Authors: | Smith, W.J, Nassar, N, Bretscher, A.P, Cerione, R.A, Karplus, P.A. | | Deposit date: | 2002-12-20 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active N-terminal Domain of Ezrin. CONFORMATIONAL AND MOBILITY CHANGES IDENTIFY KEYSTONE INTERACTIONS.
J.Biol.Chem., 278, 2003
|
|
6D2Q
 
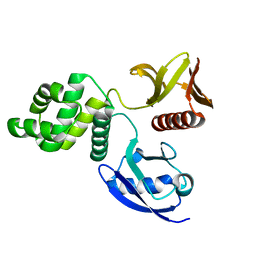 | | Crystal structure of the FERM domain of zebrafish FARP1 | | Descriptor: | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | | Authors: | Kuo, Y.C, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
6D2K
 
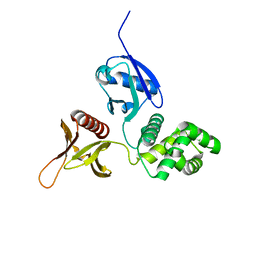 | | Crystal structure of the FERM domain of mouse FARP2 | | Descriptor: | FERM, ARHGEF and pleckstrin domain-containing protein 2 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
1EF1
 
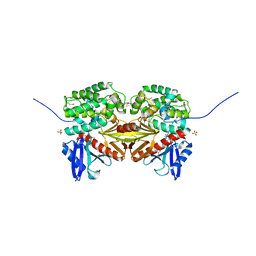 | | CRYSTAL STRUCTURE OF THE MOESIN FERM DOMAIN/TAIL DOMAIN COMPLEX | | Descriptor: | MOESIN, SULFATE ION | | Authors: | Pearson, M.A, Reczek, D, Bretscher, A, Karplus, P.A. | | Deposit date: | 2000-02-04 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain.
Cell(Cambridge,Mass.), 101, 2000
|
|
1E5W
 
 | |
6CB0
 
 | | Crystal Structure of the FAK FERM domain | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Dementiev, A, Marlowe, T. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | High resolution crystal structure of the FAK FERM domain reveals new insights on the Druggability of tyrosine 397 and the Src SH3 binding site.
BMC Mol Cell Biol, 20, 2019
|
|
6D21
 
 | |
2EMS
 
 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule CD43 | | Descriptor: | Leukosialin, Radixin | | Authors: | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the cytoplasmic tail of adhesion molecule CD43 and its binding to ERM proteins
J.Mol.Biol., 381, 2008
|
|
6CDS
 
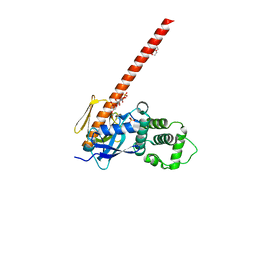 | | Human neurofibromin 2/merlin/schwannomin residues 1-339 in complex with PIP2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Merlin, ... | | Authors: | Chinthalapudi, K, Sharff, A.J, Bricogne, G, Izard, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Lipid binding promotes the open conformation and tumor-suppressive activity of neurofibromin 2.
Nat Commun, 9, 2018
|
|
2EMT
 
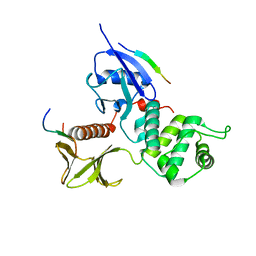 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1 | | Descriptor: | P-selectin glycoprotein ligand 1, Radixin | | Authors: | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of PSGL-1 binding to ERM proteins
Genes Cells, 12, 2007
|
|
1MK7
 
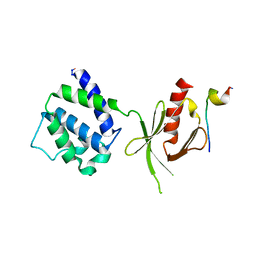 | | CRYSTAL STRUCTURE OF AN INTEGRIN BETA3-TALIN CHIMERA | | Descriptor: | Integrin Beta3, TALIN | | Authors: | Garcia-Alvarez, B, De Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinants of Integrin Recognition by Talin
Mol.Cell, 11, 2003
|
|
1MIX
 
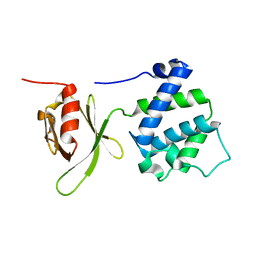 | | Crystal structure of a FERM domain of Talin | | Descriptor: | Talin | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
1MIZ
 
 | | Crystal structure of an integrin beta3-talin chimera | | Descriptor: | TALIN, integrin beta3 | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
1GG3
 
 | |
1GC6
 
 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN COMPLEXED WITH INOSITOL-(1,4,5)-TRIPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, RADIXIN | | Authors: | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2000-07-21 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
1GC7
 
 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN | | Descriptor: | RADIXIN | | Authors: | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2000-07-21 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
1MK9
 
 | | CRYSTAL STRUCTURE OF AN INTEGRIN BETA3-TALIN CHIMERA | | Descriptor: | Integrin Beta3, TALIN | | Authors: | Garcia-Alvarez, B, De Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Determinants of Integrin Recognition by Talin
Mol.Cell, 11, 2003
|
|
2D2Q
 
 | |
2YVC
 
 | |
8TEC
 
 | |
8T0D
 
 | |
