7Y02
 
 | | Crystal structure of Ricin A chain bound with (S)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-3-(4-fluorophenyl)propanoic acid | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-fluorophenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
4GUW
 
 | | Crystal structure of type 1 Ribosome inactivating protein from Momordica balsamina with lipopolysaccharide at 1.6 Angstrom resolution | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singh, A, Pandey, S, Kushwaha, G.S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of type 1 Ribosome inactivating protein from Momordica balsamina with lipopolysaccharide at 1.6 Angstrom resolution
To be published
|
|
2B7U
 
 | |
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
1LLN
 
 | |
3KTZ
 
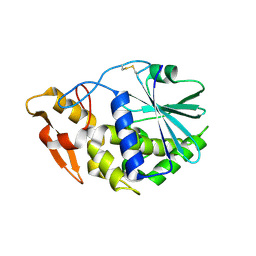 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
4DWM
 
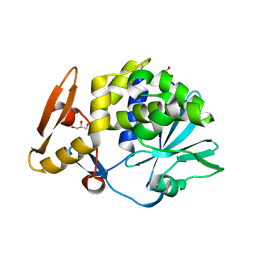 | | Crystal structure of the complex of type I Ribosome inactivating protein with N-acetylglucosamine at 1.62 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-02-25 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein with N-acetylglucosamine at 1.62 A resolution
To be Published
|
|
4KMK
 
 | | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution
To be Published
|
|
4LGR
 
 | | Ricin A chain bound to camelid nanobody (VHH3) | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M, Burshteyn, F, Cassidy, M, Gary, E, Mantis, N. | | Deposit date: | 2013-06-28 | | Release date: | 2014-06-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
J.Mol.Biol., 426, 2014
|
|
7MLO
 
 | | Crystal structure of ricin A chain in complex with 5-mesitylthiophene-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-(2,4,6-trimethylphenyl)thiophene-2-carboxylic acid, Ricin, ... | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-28 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
5Z3I
 
 | |
1LP8
 
 | |
3LE7
 
 | | Crystal structure of PD-L1 from P. dioica in complex with adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein PD-L1/PD-L2 | | Authors: | Ruggiero, A, Berisio, R. | | Deposit date: | 2010-01-14 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The role of the glycan moiety on the structure-function relationships of PD-L1, type 1 ribosome-inactivating protein from P. dioica leaves
Mol Biosyst, 6, 2010
|
|
3S9Q
 
 | | Crystal structure of native type 1 ribosome inactivating protein from Momordica balsamina at 1.67 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-06-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states.
Biochim.Biophys.Acta, 1824, 2012
|
|
4ZA3
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-13 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4LT4
 
 | | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution
To be Published
|
|
4HUP
 
 | | Structure of ricin A chain bound with N-(N-(pterin-7-yl)carbonylglycyl)-L-phenylalanyl)-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-1H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-3-phenyl-propanoic acid, MALONIC ACID, Ricin, ... | | Authors: | Jasheway, K.R, Monzingo, A.F, Saito, R, Pruet, J.M, Manzano, L.A, Wiget, P.A, Anslyn, E.V, Robertus, J.D. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Peptide-conjugated pterins as inhibitors of ricin toxin A.
J.Med.Chem., 56, 2013
|
|
4FBC
 
 | | Structure of mutant RIP from barley seeds in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1TCS
 
 | |
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
4L66
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with highly ordered water structure in the substrate binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-06-12 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with highly ordered water structure in the substrate binding site
To be Published
|
|
3MRW
 
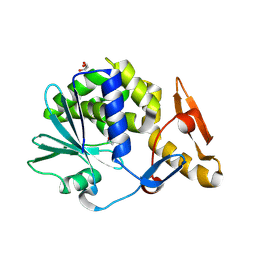 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution
To be Published
|
|
4M5A
 
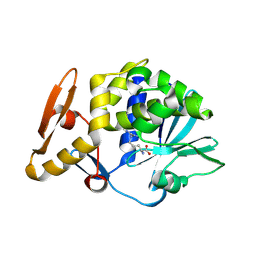 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina inhibited by asymmetric dimethyl arginine at 1.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NG,NG-DIMETHYL-L-ARGININE, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-08-08 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina inhibited by asymmetric dimethyl arginine at 1.70 A resolution
To be Published
|
|
4FXA
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl arginine at 1.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, N-ALPHA-L-ACETYL-ARGININE, ... | | Authors: | Singh, A, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-07-03 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl arginine at 1.7 Angstrom resolution
To be Published
|
|
3N3X
 
 | | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Vikram, G, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution
To be Published
|
|
