9JDV
 
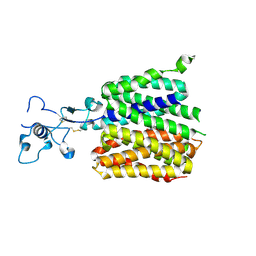 | | Human URAT1 bound with Uric acid | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Wu, C, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JDY
 
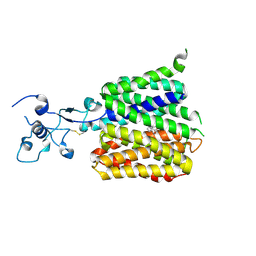 | | Human URAT1 bound with verinurad | | Descriptor: | Solute carrier family 22 member 12, verinurad | | Authors: | Wu, C, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
8WQZ
 
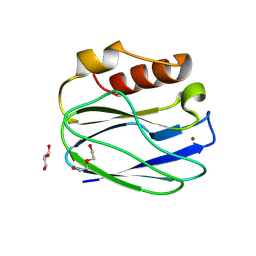 | |
9C88
 
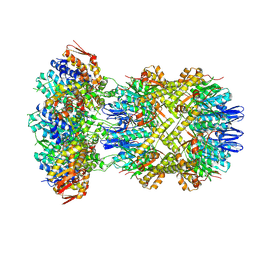 | |
8PVU
 
 | |
8RCY
 
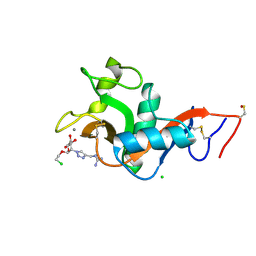 | | L-SIGN CRD in complex with Man84. | | Descriptor: | 1-[[1-[(2S,3S,4R,5S,6R)-2-(2-chloroethyloxy)-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]-1,2,3-triazol-4-yl]methyl]guanidine, C-type lectin domain family 4 member M, CALCIUM ION, ... | | Authors: | Thepaut, M, Bouchikri, C, Pollastri, S, Bernardi, A, Fieschi, F. | | Deposit date: | 2023-12-07 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented selectivity for homologous lectin targets: differential targeting of the viral receptors L-SIGN and DC-SIGN.
Chem Sci, 15, 2024
|
|
8RLU
 
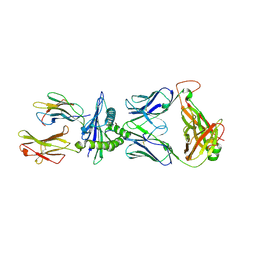 | |
9BA1
 
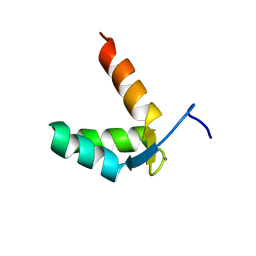 | |
9D7K
 
 | | Infectious B19V capsid | | Descriptor: | Alpha-1-antichymotrypsin | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Infectious parvovirus B19 circulates in the blood plasma coated with active host protease inhibitors
To Be Published
|
|
9FMR
 
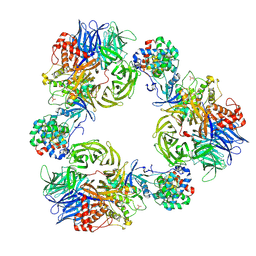 | |
8WPN
 
 | |
9BI9
 
 | |
9C27
 
 | | Infectious B19V capsid | | Descriptor: | Capsid protein 2 | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Infectious parvovirus B19 circulates in the blood plasma coated with active host protease inhibitors
To Be Published
|
|
8Q4Y
 
 | | Beta-galactosidase from Bacillus circulans conformational state 1 | | Descriptor: | beta-galactosidase | | Authors: | Kascakova, B, Hovorkova, M, Petraskova, L, Novacek, J, Pinkas, D, Gardian, Z, Kren, V, Bojarova, P, Kuta Smatanova, I. | | Deposit date: | 2023-08-07 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The variable structural flexibility of the Bacillus circulans beta-galactosidase isoforms determines their unique functionalities.
Structure, 2024
|
|
8QDN
 
 | |
8QE6
 
 | |
8UEN
 
 | |
8UGM
 
 | |
8QMV
 
 | | L2 forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of Form I'' and Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Layered entrenchment maintains essentiality in the evolution of Form I Rubisco complexes
Biorxiv, 2024
|
|
8QPS
 
 | |
8QQN
 
 | |
8R6X
 
 | |
8RDO
 
 | |
8RDL
 
 | | Xenorhabdin methyltransferase XrdM with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ToxA protein | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isofunctional but Structurally Different Methyltransferases for Dithiolopyrrolone Diversification.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8RDM
 
 | | Holomycin methyltransferase DtpM with SAH | | Descriptor: | DtpM, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isofunctional but Structurally Different Methyltransferases for Dithiolopyrrolone Diversification.
Angew.Chem.Int.Ed.Engl., 2024
|
|
