8PPL
 
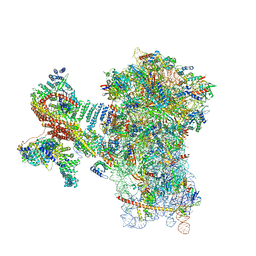 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
3CFP
 
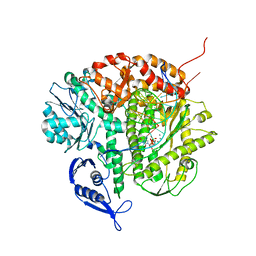 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
3CFR
 
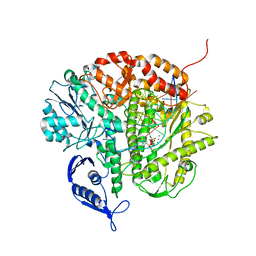 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*(DOC))-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
6O81
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B bound to translation initiation factor 2 from Homo sapiens | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Nguyen, H, Kenner, L, Frost, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response.
Science, 364, 2019
|
|
4V4B
 
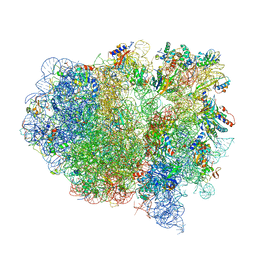 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
3K59
 
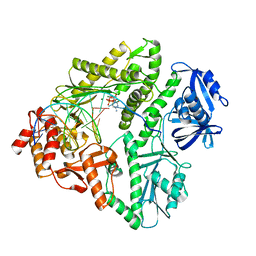 | | Crystal structure of E.coli Pol II-normal DNA-dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*TP*GP*CP*CP*TP*AP*GP*CP*GP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*AP*GP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II
Cell(Cambridge,Mass.), 139, 2009
|
|
3K5N
 
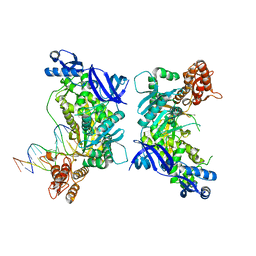 | | Crystal structure of E.coli Pol II-abasic DNA binary complex | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*TP*GP*(3DR)*TP*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*TP*AP*GP*CP*GP*TP*AP*G)-3'), DNA polymerase II | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II.
Cell(Cambridge,Mass.), 139, 2009
|
|
3K5M
 
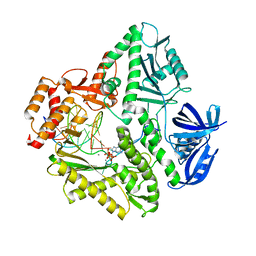 | | Crystal structure of E.coli Pol II-abasic DNA-ddGTP Lt(-2, 2) ternary complex | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*TP*CP*CP*TP*GP*(3DR)P*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II.
Cell(Cambridge,Mass.), 139, 2009
|
|
6OAR
 
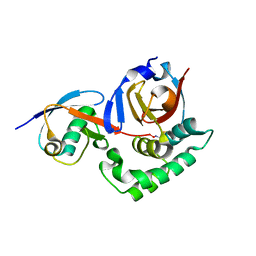 | |
6OAT
 
 | | Structure of the Ganjam virus OTU bound to sheep ISG15 | | Descriptor: | Interferon stimulated gene 17, RNA-dependent RNA polymerase, prop-2-en-1-amine | | Authors: | Dzimianski, J.V, Williams, I.L, Pegan, S.D. | | Deposit date: | 2019-03-18 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Determining the molecular drivers of species-specific interferon-stimulated gene product 15 interactions with nairovirus ovarian tumor domain proteases.
Plos One, 14, 2019
|
|
1QFG
 
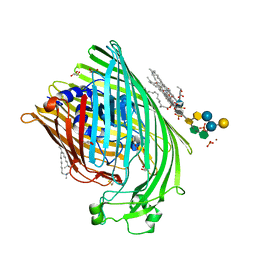 | | E. COLI FERRIC HYDROXAMATE RECEPTOR (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Welte, W, Hofmann, E, Lindner, B, Holst, O, Coulton, J.W, Diederichs, K. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
1GPA
 
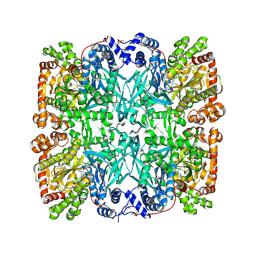 | |
6N8O
 
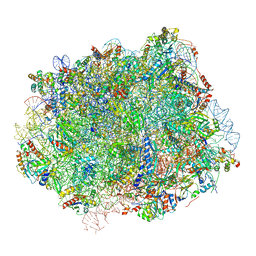 | | Cryo-EM structure of Rpl10-inserted (RI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8J
 
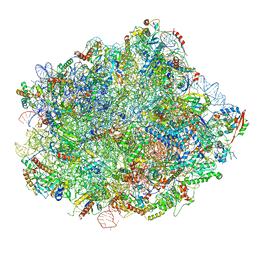 | | Cryo-EM structure of late nuclear (LN) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal protein L11-A, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8L
 
 | | Cryo-EM structure of early cytoplasmic-late (ECL) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8M
 
 | | Cryo-EM structure of pre-Lsg1 (PL) pre-60S ribosomal subunit | | Descriptor: | 5.8S RNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8N
 
 | | Cryo-EM structure of Lsg1-engaged (LE) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
5LZU
 
 | | Structure of the mammalian ribosomal termination complex with accommodated eRF1 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
6W2S
 
 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the open state (Class 1) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
1HAA
 
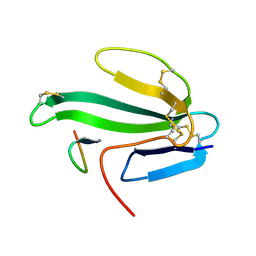 | | A beta-Hairpin Structure in a 13-mer Peptide that Binds a-Bungarotoxin with High Affinity and Neutralizes its Toxicity | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE | | Authors: | Scherf, T, Kasher, R, Balass, M, Fridkin, M, Fuchs, S, Katchalski-Katzir, E. | | Deposit date: | 2001-04-05 | | Release date: | 2001-05-25 | | Last modified: | 2017-02-08 | | Method: | SOLUTION NMR | | Cite: | A Beta-Hairpin Structure in a 13-mer Peptide that Binds Alpha-Bungarotoxin with High Affinity and Neutralizes its Toxicity
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3D3I
 
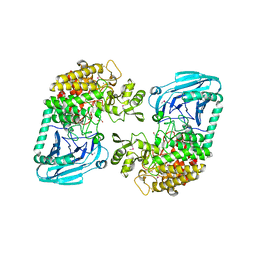 | | Crystal structural of Escherichia coli K12 YgjK, a glucosidase belonging to glycoside hydrolase family 63 | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein ygjK | | Authors: | Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2008-05-12 | | Release date: | 2008-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
1QSP
 
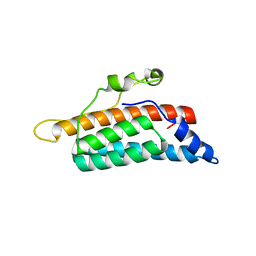 | | CRYSTAL STRUCTURE OF THE YEAST PHOSPHORELAY PROTEIN YPD1 | | Descriptor: | YPD1 | | Authors: | Xu, Q, West, A.H. | | Deposit date: | 1999-06-22 | | Release date: | 1999-10-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1.
J.Mol.Biol., 292, 1999
|
|
1QSY
 
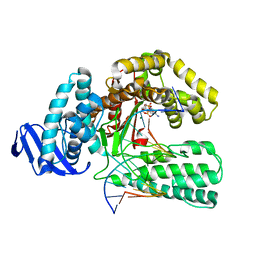 | | DDATP-Trapped closed ternary complex of the large fragment of DNA Polymerase I from thermus aquaticus | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, 5'-D(*AP*TP*TP*GP*CP*GP*CP*CP*TP*P*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3', ... | | Authors: | Li, Y, Mitaxov, V, Waksman, G. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1HE5
 
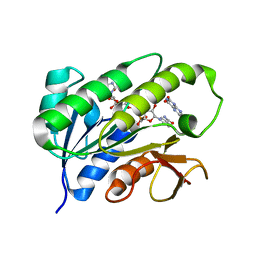 | | Human biliverdin IX beta reductase: NADP/Lumichrome ternary complex | | Descriptor: | BILIVERDIN IX BETA REDUCTASE, LUMICHROME, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pereira, P.J.B, Macedo-Ribeiro, S, Parraga, A, Perez-Luque, R, Cunningham, O, Darcy, K, Mantle, T.J, Coll, M. | | Deposit date: | 2000-11-19 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Human Biliverdin Ix Beta Reductase, an Early Fetal Bilirubin Ix Producing Enzyme
Nat.Struct.Biol., 8, 2001
|
|
1HE4
 
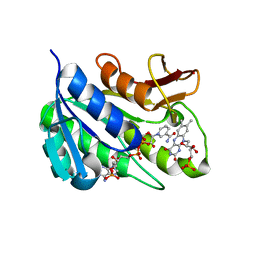 | | Human biliverdin IX beta reductase: NADP/FMN ternary complex | | Descriptor: | BILIVERDIN IX BETA REDUCTASE, FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pereira, P.J.B, Macedo-Ribeiro, S, Parraga, A, Perez-Luque, R, Cunningham, O, Darcy, K, Mantle, T.J, Coll, M. | | Deposit date: | 2000-11-19 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Human Biliverdin Ix Beta Reductase, an Early Fetal Bilirubin Ix Producing Enzyme
Nat.Struct.Biol., 8, 2001
|
|
