2PE4
 
 | | Structure of Human Hyaluronidase 1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Chao, K.L, Herzberg, O. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human Hyaluronidase-1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis
Biochemistry, 46, 2007
|
|
4ZOH
 
 | | Crystal structure of glyceraldehyde oxidoreductase | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nishimasu, H, Fushinobu, S, Wakagi, T. | | Deposit date: | 2015-05-06 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Archaeal Mo-Containing Glyceraldehyde Oxidoreductase Isozymes Exhibit Diverse Substrate Specificities through Unique Subunit Assemblies.
Plos One, 11, 2016
|
|
3H3D
 
 | |
5ZJT
 
 | | Structure of AbdB/Exd complex bound to a 'Black14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*AP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*TP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Zeiske, T, Baburajendran, N, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5ZJS
 
 | | Structure of AbdB/Exd complex bound to a 'Blue14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*TP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*AP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Baburajendran, N, Zeiske, T, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
4XGC
 
 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
4XB7
 
 | | Crystal structure of Dscam1 isoform 4.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.4, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4JBK
 
 | | Molecular basis for abrogation of activation of pro-inflammatory cytokines | | Descriptor: | DNA (5'-D(P*GP*GP*AP*AP*TP*TP*AP*TP*AP*AP*TP*TP*CP*C)-3'), Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4YVJ
 
 | |
1KRR
 
 | |
1KRV
 
 | |
5VKQ
 
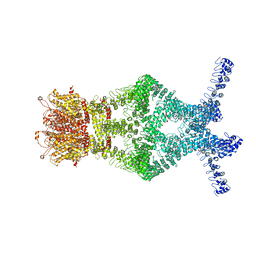 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | Authors: | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | Deposit date: | 2017-04-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
5W76
 
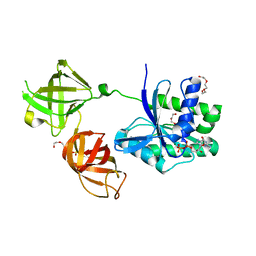 | | Crystal Structure of Reconstructed Bacterial Elongation Factor Node 168 | | Descriptor: | Ancestral Elogation Factor N153, DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ortlund, E.A. | | Deposit date: | 2017-06-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural and Dynamics Comparison of Thermostability in Ancient, Modern, and Consensus Elongation Factor Tus.
Structure, 26, 2018
|
|
5H7V
 
 | |
5WJE
 
 | | Crystal structure of Naa80 bound to a bisubstrate analogue | | Descriptor: | Actin N-terminus peptide, CARBOXYMETHYL COENZYME *A, CG8481, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5H9B
 
 | |
3GPX
 
 | | Sequence-matched MutM Interrogation Complex 4 (IC4) | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*AP*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Spong, M.C, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
3GQ5
 
 | | Sequence-matched MutM Interrogation Complex 5 (IC5) | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*CP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*T*CP*CP*GP*GP*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
5Z4J
 
 | | Structure of Tailor in complex with U4 RNA | | Descriptor: | RNA (5'-R(*UP*UP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5HLE
 
 | |
5Y3J
 
 | | Crystal structure of horse TLR9 in complex with two DNAs (CpG DNA and TCGCAC DNA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2017-07-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Immunity, 48, 2018
|
|
5Y3K
 
 | | Crystal structure of horse TLR9 in complex with two DNAs (CpG DNA and GCGCAC DNA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2017-07-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Immunity, 48, 2018
|
|
4I0P
 
 | | HLA-DO in complex with HLA-DM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Guce, A.I, Mortimer, S.E, Stern, L.J. | | Deposit date: | 2012-11-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5X2P
 
 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
2Q86
 
 | | Structure of the mouse invariant NKT cell receptor Valpha14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Valpha14 TCR, Vbeta8.2, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2007-06-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Mouse CD1d-iGb3 Complex and its Cognate Valpha14 T Cell Receptor Suggest a Model for Dual Recognition of Foreign and Self Glycolipids.
J.Mol.Biol., 377, 2008
|
|
