3WYM
 
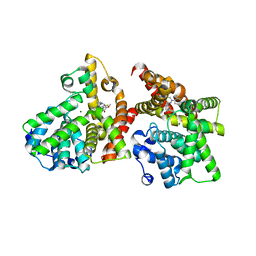 | | Crystal structure of the catalytic domain of PDE10A complexed with 1-(2-fluoro-4-(1H-pyrazol-1-yl)phenyl)-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Hayano, Y. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 1-[2-fluoro-4-(1H-pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor.
J.Med.Chem., 57, 2014
|
|
1I2Z
 
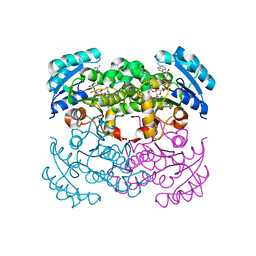 | | E. COLI ENOYL REDUCTASE IN COMPLEX WITH NAD AND BRL-12654 | | Descriptor: | 4-(2-THIENYL)-1-(4-METHYLBENZYL)-1H-IMIDAZOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Heerding, D.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,4-Disubstituted imidazoles are potential antibacterial agents functioning as inhibitors of enoyl acyl carrier protein
reductase (FabI).
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1R0L
 
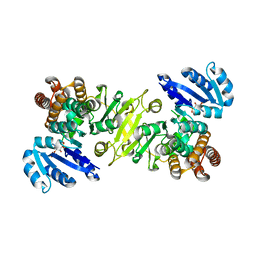 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from zymomonas mobilis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
1QS4
 
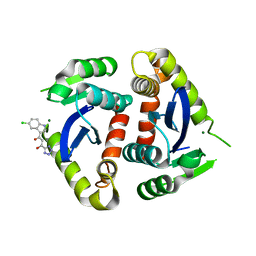 | | Core domain of HIV-1 integrase complexed with Mg++ and 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone | | Descriptor: | 1-(5-CHLOROINDOL-3-YL)-3-HYDROXY-3-(2H-TETRAZOL-5-YL)-PROPENONE, MAGNESIUM ION, PROTEIN (HIV-1 INTEGRASE (E.C.2.7.7.49)) | | Authors: | Goldgur, Y, Craigie, R, Fujiwara, T, Yoshinaga, T, Davies, D.R. | | Deposit date: | 1999-06-25 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3PMO
 
 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2QUU
 
 | |
1FO2
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE IN COMPLEX WITH 1-DEOXYMANNOJIRIMYCIN | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, ALPHA1,2-MANNOSIDASE, CALCIUM ION, ... | | Authors: | Vallee, F, Karaveg, K, Moremen, K.W, Herscovics, A, Howell, P.L. | | Deposit date: | 2000-08-24 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
1RTI
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 1-(2-HYDROXYETHYLOXYMETHYL)-6-PHENYL THIOTHYMINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1CRA
 
 | | THE COMPLEX BETWEEN HUMAN CARBONIC ANHYDRASE II AND THE AROMATIC INHIBITOR 1,2,4-TRIAZOLE | | Descriptor: | 1,2,4-TRIAZOLE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Mangani, S, Liljas, A. | | Deposit date: | 1992-10-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex between human carbonic anhydrase II and the aromatic inhibitor 1,2,4-triazole.
J.Mol.Biol., 232, 1993
|
|
3TNL
 
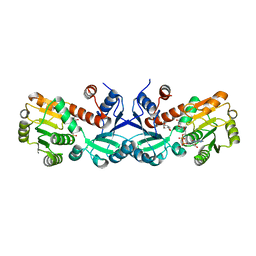 | | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD. | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD.
TO BE PUBLISHED
|
|
1KQP
 
 | | NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS AT 1 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Symersky, J, Devedjiev, Y, Moore, K, Brouillette, C, DeLucas, L. | | Deposit date: | 2002-01-07 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | NH3-dependent NAD+ synthetase from Bacillus subtilis at 1 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3TOF
 
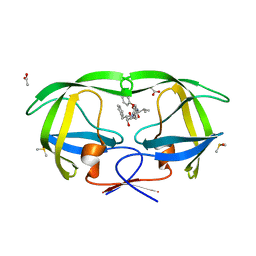 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 6 - Orthorombic Crystal form P212121) | | Descriptor: | (S)-N-((1R,2S)-1-((2R,3R)-3-benzyloxiran-2-yl)-1-hydroxy-3-phenylpropan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Geremia, S, Olajuyigbe, F.M, Ajele, J.O, Demitri, N, Randaccio, L, Wuerges, J, Benedetti, L, Campaner, P, Berti, F. | | Deposit date: | 2011-09-05 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
3TOG
 
 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 9 - Monoclinic Crystal form P21) | | Descriptor: | (S)-N-((2S,3S,4R,5R)-4-amino-3,5-dihydroxy-1,6-diphenylhexan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, DIMETHYL SULFOXIDE, Gag-Pol polyprotein | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2011-09-05 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
3TOH
 
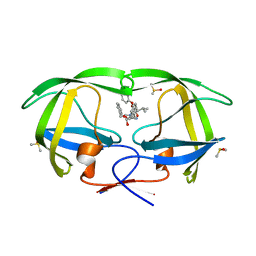 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 9 - Orthorombic Crystal form P212121) | | Descriptor: | (S)-N-((2S,3S,4R,5R)-4-amino-3,5-dihydroxy-1,6-diphenylhexan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, DIMETHYL SULFOXIDE, Gag-Pol polyprotein | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2011-09-05 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.116 Å) | | Cite: | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
2J8F
 
 | |
2QUT
 
 | |
2LYA
 
 | |
2JPR
 
 | | Joint refinement of the HIV-1 CA-NTD in complex with the assembly inhibitor CAP-1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, Gag-Pol polyprotein | | Authors: | Kelly, B.N, Kyere, S, Kinde, I, Tang, C, Howard, B.R, Robinson, H, Sundquist, W.I, Summers, M.F, Hill, C.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein
J.Mol.Biol., 373, 2007
|
|
2FZZ
 
 | | Factor Xa in complex with the inhibitor 1-(3-amino-1,2-benzisoxazol-5-yl)-6-(2'-(((3r)-3-hydroxy-1-pyrrolidinyl)methyl)-4-biphenylyl)-3-(trifluoromethyl)-1,4,5,6-tetrahydro-7h-pyrazolo[3,4-c]pyridin-7-one | | Descriptor: | 1-(3-AMINO-1,2-BENZISOXAZOL-5-YL)-6-(2'-{[(3R)-3-HYDROXYPYRROLIDIN-1-YL]METHYL}BIPHENYL-4-YL)-3-(TRIFLUOROMETHYL)-1,4,5,6-TETRAHYDRO-7H-PYRAZOLO[3,4-C]PYRIDIN-7-ONE, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2006-02-10 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1-[3-Aminobenzisoxazol-5'-yl]-3-trifluoromethyl-6-[2'-(3-(R)-hydroxy-N-pyrrolidinyl)methyl-[1,1']-biphen-4-yl]-1,4,5,6-tetrahydropyrazolo-[3,4-c]-pyridin-7-one (BMS-740808) a highly potent, selective, efficacious, and orally bioavailable inhibitor of blood coagulation factor Xa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FJK
 
 | | Crystal structure of Fructose-1,6-Bisphosphate Aldolase in Thermus caldophilus | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase | | Authors: | Lee, J.H, Im, Y.J, Rho, S.-H, Kim, M.-K, Kang, G.B, Eom, S.H. | | Deposit date: | 2006-01-03 | | Release date: | 2006-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stereoselectivity of fructose-1,6-bisphosphate aldolase in Thermus caldophilus
Biochem.Biophys.Res.Commun., 347, 2006
|
|
6CR2
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from Aspergillus fumigatus in complex with the VNI derivative N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(2-fluoro-4-(2,2,2-trifluoroethoxy)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | 14-alpha sterol demethylase Cyp51B, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[2-fluoro-4-(2,2,2-trifluoroethoxy)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2018-03-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Sterol 14 alpha-Demethylase Structure-Based Design of VNI (( R)- N-(1-(2,4-Dichlorophenyl)-2-(1 H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide)) Derivatives To Target Fungal Infections: Synthesis, Biological Evaluation, and Crystallographic Analysis.
J. Med. Chem., 61, 2018
|
|
1LUC
 
 | | BACTERIAL LUCIFERASE | | Descriptor: | 1,2-ETHANEDIOL, BACTERIAL LUCIFERASE, MAGNESIUM ION | | Authors: | Fisher, A.J, Rayment, I. | | Deposit date: | 1996-05-10 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5-A resolution crystal structure of bacterial luciferase in low salt conditions.
J.Biol.Chem., 271, 1996
|
|
4A27
 
 | | Crystal structure of human synaptic vesicle membrane protein VAT-1 homolog-like protein | | Descriptor: | 1,2-ETHANEDIOL, SYNAPTIC VESICLE MEMBRANE PROTEIN VAT-1 HOMOLOG-LIKE | | Authors: | Vollmar, M, Shafqat, N, Muniz, J.R.C, Krojer, T, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Synaptic Vesicle Membrane Protein Vat-1 Homolog-Like Protein
To be Published
|
|
6PZ1
 
 | | Crystal Structure of human Indoleamine 2,3-Dioxygenase 1 in complex with PF-06840003 in Active Site and Si site | | Descriptor: | (3R)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Pham, K.N, Lewis-Ballester, A, Yeh, S.R. | | Deposit date: | 2019-07-31 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in Human Indoleamine 2,3-Dioxygenase 1 and Tryptophan Dioxygenase.
J.Am.Chem.Soc., 141, 2019
|
|
4UB8
 
 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
