4L6Q
 
 | | ROCK2 in complex with benzoxaborole | | Descriptor: | 6-[4-(aminomethyl)-2-fluorophenoxy]-2,1-benzoxaborol-1(3H)-ol, Rho-associated protein kinase 2 | | Authors: | Rock, F, Jarnagin, K. | | Deposit date: | 2013-06-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Linking phenotype to kinase: identification of a novel benzoxaborole hinge-binding motif for kinase inhibition and development of high-potency rho kinase inhibitors.
J.Pharmacol.Exp.Ther., 347, 2013
|
|
1Y9T
 
 | |
3AX8
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 15alpha-methoxy-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3S,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-3-methoxy-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-03-30 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New C15-substituted active vitamin D3
Org.Lett., 13, 2011
|
|
4AE4
 
 | | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a novel SOUBA domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Agromayor, M, Soler, N, Caballe, A, Kueck, T, Freund, S.M, Allen, M.D, Bycroft, M, Perisic, O, Ye, Y, McDonald, B, Scheel, H, Hofmann, K, Neil, S.J.D, Martin-Serrano, J, Williams, R.L. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain.
Structure, 20, 2012
|
|
1U8A
 
 | | Crystal Structure of Mycobacterium Tuberculosis Shikimate Kinase in Complex with Shikimate and ADP at 2.15 Angstrom Resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Dhaliwal, B, Nichols, C.E, Ren, J, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-05 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic studies of shikimate binding and induced conformational changes in Mycobacterium tuberculosis shikimate kinase.
Febs Lett., 574, 2004
|
|
1KHI
 
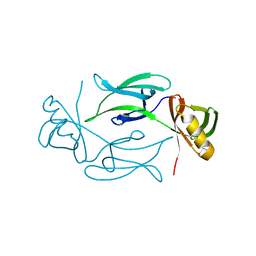 | | CRYSTAL STRUCTURE OF HEX1 | | Descriptor: | Hex1 | | Authors: | Yuan, P, Swaminathan, K. | | Deposit date: | 2001-11-30 | | Release date: | 2002-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa
NAT.STRUCT.BIOL., 10, 2003
|
|
3AXK
 
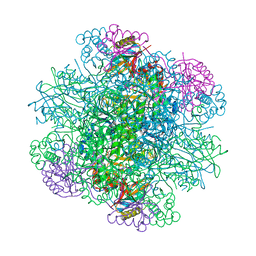 | | Structure of rice Rubisco in complex with NADP(H) | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
6B1F
 
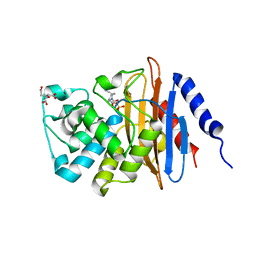 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 4234 by soaking | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
4CFD
 
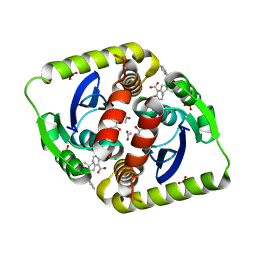 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 5-[[[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-phenyl-ethyl]carbamoyl-methyl-amino]methyl]-1,3-benzodioxole-4-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CE9
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-2-(2-carboxyethyl)-6-{[{2-[(cyclohexylmethyl)carbamoyl]benzyl}(prop-2-en-1-yl)amino]methyl}-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
7HET
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003693 | | Descriptor: | (2R)-2-[(1R)-2,3-dihydro-1H-inden-1-yl]-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]ethan-1-ol, (2S)-2-[(1R)-2,3-dihydro-1H-inden-1-yl]-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]ethan-1-ol, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
4JKI
 
 | | Crystal Structure of N10-Formyltetrahydrofolate Synthetase with ZD9331, Formylphosphate, and ADP | | Descriptor: | 2,7-dimethyl-6-[(prop-1-yn-1-ylamino)methyl]quinazolin-4(3H)-one, ADENOSINE-5'-DIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Celeste, L.R, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2013-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of N10-formyltetrahydrofolate synthetase derived from complexes with intermediates and inhibitors.
Protein Sci., 21, 2012
|
|
6ATF
 
 | | HLA-DRB1*1402 in complex with Vimentin59-71 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scally, S.W, Ting, Y.T, Rossjohn, J. | | Deposit date: | 2017-08-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for increased susceptibility of Indigenous North Americans to seropositive rheumatoid arthritis.
Ann. Rheum. Dis., 76, 2017
|
|
3NM5
 
 | | Helicobacter pylori MTAN complexed with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Ronning, D.R, Iacopelli, N.M. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
Protein Sci., 19, 2010
|
|
2R5S
 
 | | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Wu, R, Abdullah, J, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
6B1H
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 4234 by co-crystallization | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, ACETATE ION, CHLORIDE ION, ... | | Authors: | van den Akker, F, Nhuyen, N.Q. | | Deposit date: | 2017-09-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
7HTZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z69092635 | | Descriptor: | N-[2-(cyclohex-1-en-1-yl)ethyl]-3,5-dimethyl-1,2-oxazole-4-carboxamide, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic, ... | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3D2L
 
 | |
3NAY
 
 | | PDK1 in complex with inhibitor MP6 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-(2-cyclopropylethylidene)-9-(1H-pyrazol-4-yl)-6-{[(1R)-1,2,2-trimethylpropyl]amino}benzo[c][1,6]naphthyridin-1(4H)-one | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective inhibition of PDK1 using an allosteric kinase inhibitor and RNAi impairs cancer cell migration and anchorage-independent growth of primary tumor lines
J.Biol.Chem., 2010
|
|
3D33
 
 | |
3S7B
 
 | | Structural Basis of Substrate Methylation and Inhibition of SMYD2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[2-(5-hydroxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2011-05-26 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Methylation and Inhibition of SMYD2.
Structure, 19, 2011
|
|
1T4P
 
 | | Arginase-dehydro-ABH complex | | Descriptor: | Arginase 1, MANGANESE (II) ION, [(1E,5S)-5-AMINO-5-CARBOXYPENT-1-ENYL](TRIHYDROXY)BORATE(1-) | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Han, S, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1I1O
 
 | |
1DYS
 
 | | Endoglucanase CEL6B from Humicola insolens | | Descriptor: | ENDOGLUCANASE | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, M, Varrot, A, Schulein, M. | | Deposit date: | 2000-02-08 | | Release date: | 2001-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Humicola Insolens Family 6 Cellulases: Structure of the Endoglucanase, Cel6B, at 1.6 A Resolution
Biochem.J., 348, 2000
|
|
6B88
 
 | | E. coli LepB in complex with GNE0775 ((4S,7S,10S)-10-((S)-4-amino-2-(2-(4-(tert-butyl)phenyl)-4-methylpyrimidine-5-carboxamido)-N-methylbutanamido)-16,26-bis(2-aminoethoxy)-N-(2-iminoethyl)-7-methyl-6,9-dioxo-5,8-diaza-1,2(1,3)-dibenzenacyclodecaphane-4-carboxamide) | | Descriptor: | (8S,11S,14S)-14-{[(2S)-4-amino-2-{[2-(4-tert-butylphenyl)-4-methylpyrimidine-5-carbonyl]amino}butanoyl](methyl)amino}-3,18-bis(2-aminoethoxy)-N-[(2Z)-2-iminoethyl]-11-methyl-10,13-dioxo-9,12-diazatricyclo[13.3.1.1~2,6~]icosa-1(19),2(20),3,5,15,17-hexaene-8-carboxamide, PENTAETHYLENE GLYCOL, Signal peptidase I | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Optimized arylomycins are a new class of Gram-negative antibiotics.
Nature, 561, 2018
|
|
