6YWE
 
 | | The structure of the mitoribosome from Neurospora crassa in the P/E tRNA bound state | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
5MCW
 
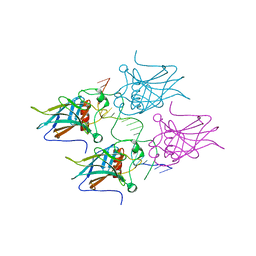 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC2) | | Descriptor: | Cellular tumor antigen p53, DNA, FORMYL GROUP, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
2O02
 
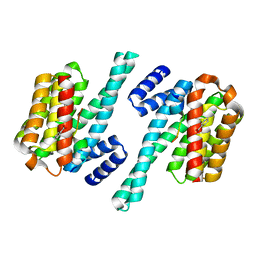 | | Phosphorylation independent interactions between 14-3-3 and Exoenzyme S: from structure to pathogenesis | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, ExoS (416-430) peptide | | Authors: | Ottmann, C, Yasmin, L, Weyand, M, Hauser, A.R, Wittinghofer, A, Hallberg, B. | | Deposit date: | 2006-11-27 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: from structure to pathogenesis
Embo J., 26, 2007
|
|
7T2Y
 
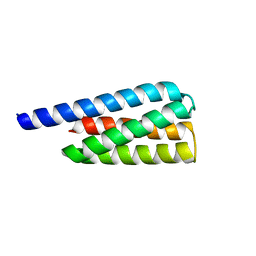 | | X-ray structure of a designed cold unfolding four helix bundle | | Descriptor: | Designed cold unfolding four helix bundle | | Authors: | Harrison, J.S, Kuhlman, B, Szyperski, T, Premkumar, L, Maguire, J, Pulavarti, S, Yuen, S. | | Deposit date: | 2021-12-06 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
5MUS
 
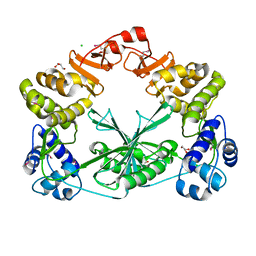 | | Structure of the C-terminal domain of a reptarenavirus L protein | | Descriptor: | CHLORIDE ION, GLYCEROL, L protein | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
1O3T
 
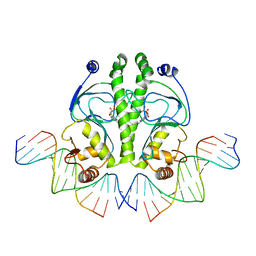 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex:
DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
6YWV
 
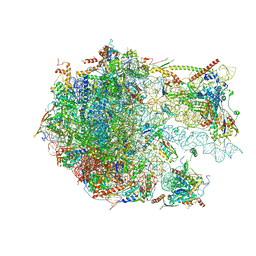 | | The structure of the Atp25 bound assembly intermediate of the mitoribosome from Neurospora crassa | | Descriptor: | 23 S rRNA, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
1A8V
 
 | |
2HHT
 
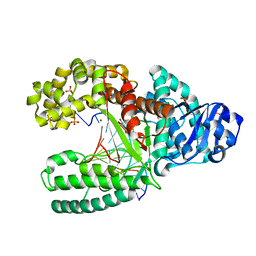 | | C:O6-methyl-guanine pair in the polymerase-2 basepair position | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*CP*G)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2VC8
 
 | |
5MDQ
 
 | |
1JHN
 
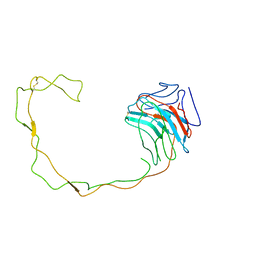 | | Crystal Structure of the Lumenal Domain of Calnexin | | Descriptor: | CALCIUM ION, calnexin | | Authors: | Schrag, J.D, Bergeron, J.M, Li, Y, Borisova, S, Hahn, M, Thomas, D.Y, Cygler, M. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of calnexin, an ER chaperone involved in quality control of protein folding.
Mol.Cell, 8, 2001
|
|
5NQR
 
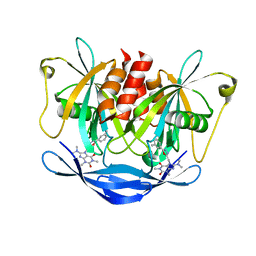 | | Potent inhibitors of NUDT5 silence hormone signaling in breast cancer | | Descriptor: | 8-(dimethylamino)-1,3-dimethyl-7-[[5-(3-methylphenyl)-1,3,4-oxadiazol-2-yl]methyl]purine-2,6-dione, ADP-sugar pyrophosphatase | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2017-04-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells.
Nat Commun, 9, 2018
|
|
8AQU
 
 | | BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQV
 
 | | BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQS
 
 | | BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
1YED
 
 | |
5ND4
 
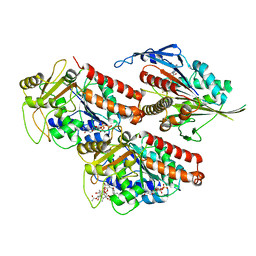 | | Microtubule-bound MKLP2 motor domain in the presence of ADP.AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Yu, I.-M, Cook, A, Muretta, J.M, Joseph, A.P, Major, J, Sourigues, Y, Clause, J, Topf, M, Rosenfeld, S.S, Houdusse, A, Moores, C.A. | | Deposit date: | 2017-03-07 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry.
Elife, 6, 2017
|
|
8PD6
 
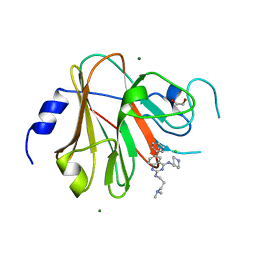 | | Crystal structure of the TRIM58 PRY-SPRY domain in complex with TRIM-473 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase TRIM58, ... | | Authors: | Renatus, M, Hoegenauer, K, Schroeder, M. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Ligands for TRIM58, a Novel Tissue-Selective E3 Ligase.
Acs Med.Chem.Lett., 14, 2023
|
|
8PD4
 
 | |
5ND2
 
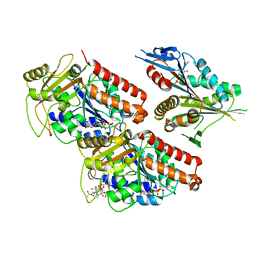 | | Microtubule-bound MKLP2 motor domain in the presence of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Yu, I.M, Cook, A, Muretta, J.M, Joseph, A.P, Major, J, Sourigues, Y, Clause, J, Topf, M, Rosenfeld, S.S, Houdusse, A, Moores, C.A. | | Deposit date: | 2017-03-07 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry.
Elife, 6, 2017
|
|
8BD2
 
 | | Calcium-bound Calmodulin variant G113R | | Descriptor: | CALCIUM ION, Calmodulin-3 | | Authors: | Wimmer, R, Holler, C.V, Petersson, N.M, Iwai, H, Niemelae, M.A, Brohus, M, Overgaard, M.T. | | Deposit date: | 2022-10-18 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Allosteric changes in protein stability and dynamics as pathogenic mechanism for calmodulin variants not affecting Ca 2+ coordinating residues.
Cell Calcium, 117, 2023
|
|
5NFP
 
 | | Glucocorticoid Receptor in complex with budesonide | | Descriptor: | (1~{S},2~{S},4~{R},6~{R},8~{S},9~{S},11~{S},12~{S},13~{R})-9,13-dimethyl-11-oxidanyl-8-(2-oxidanylethanoyl)-6-propyl-5,7-dioxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosa-14,17-dien-16-one, 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Edman, K, Wissler, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Nonsteroidal Glucocorticoid Receptor Modulators for the Inhaled Treatment of Pulmonary Diseases.
J. Med. Chem., 60, 2017
|
|
2HHQ
 
 | | O6-methyl-guanine:T pair in the polymerase-10 basepair position | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*AP*TP*GP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5NQ5
 
 | | Mtb TMK crystal structure in complex with compound 1 | | Descriptor: | 5-methyl-1-[(3~{S})-1-[(3-phenoxyphenyl)methyl]piperidin-3-yl]pyrimidine-2,4-dione, Thymidylate kinase | | Authors: | Merceron, R, Song, L, Munier-Lehmann, H, Van Calenbergh, S, Savvides, S. | | Deposit date: | 2017-04-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure Guided Lead Generation toward Nonchiral M. tuberculosis Thymidylate Kinase Inhibitors.
J. Med. Chem., 61, 2018
|
|
