6VT6
 
 | | Naegleria gruberi RNA ligase K170A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTG
 
 | | Naegleria gruberi RNA ligase E227A mutant apo | | Descriptor: | RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
4CC5
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
3UQ8
 
 | |
2E1H
 
 | |
2DJZ
 
 | |
4EQ5
 
 | | DNA ligase from the archaeon Thermococcus sibiricus | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase | | Authors: | Petrova, T, Bezsudnova, E.Y, Dorokhov, B.D, Slutskaya, E.S, Polyakov, K.M, Dorovatovskiy, P.V, Ravin, N.V, Skryabin, K.G, Kovalchuk, M.V, Popov, V.O. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Expression, purification, crystallization and preliminary crystallographic analysis of a thermostable DNA ligase from the archaeon Thermococcus sibiricus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2PVP
 
 | |
4UFZ
 
 | | Synthesis of Novel NAD Dependant DNA Ligase Inhibitors via Negishi Cross-Coupling: Development of SAR and Resistance Studies | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5,7-bis(azanyl)-2-tert-butyl-4-(1,3-thiazol-2-yl)pyrido[2,3-d]pyrimidine-6-carbonitrile, DNA LIGASE | | Authors: | Murphy-Benenato, K.E, Boriack-Sjodin, P.A, Martinez-Botella, G, Carcanague, D, Gingipali, L, Gowravaram, M, Harang, J, Hale, M, Ioannidis, G, Jahic, H, Johnstone, M, Kutschke, A, Laganas, V.A, Loch, J, Oguto, H, Patel, S.J. | | Deposit date: | 2015-03-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Negishi Cross-Coupling Enabled Synthesis of Novel Nad(+)-Dependent DNA Ligase Inhibitors and Sar Development.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5HPK
 
 | | System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes: NEDD4L and UbV NL.1 | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, Ubiquitin variant NL.1 | | Authors: | Wu, K.-P, Mukherjee, M, Mercredi, P.Y, Schulman, B.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
5BSW
 
 | |
5YWR
 
 | | Crystal Structure of RING E3 ligase ZNRF1 in complex with Ube2N (Ubc13) | | Descriptor: | E3 ubiquitin-protein ligase ZNRF1, FORMIC ACID, TRIETHYLENE GLYCOL, ... | | Authors: | Behera, A.P, Naskar, P, Datta, A.B. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural insights into the nanomolar affinity of RING E3 ligase ZNRF1 for Ube2N and its functional implications.
Biochem. J., 475, 2018
|
|
3R5X
 
 | | Crystal Structure of D-alanine--D-Alanine Ligase from Bacillus anthracis complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, Y, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-20 | | Release date: | 2011-04-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of D-alanine--D-Alanine Ligase from Bacillus anthracis complexed with ATP
To be Published
|
|
8ORJ
 
 | | Cryo-EM structure of human tRNA ligase RTCB in complex with human PYROXD1. | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Loeff, L, Kroupova, A, Asanovic, I, Boneberg, F, Pfleiderer, M.M, Ferdigg, A, Ackle, F, Martinez, J, Jinek, M. | | Deposit date: | 2023-04-14 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic basis for PYROXD1-mediated protection of the human tRNA ligase complex against oxidative inactivation.
Nat.Struct.Mol.Biol., 2025
|
|
5BSR
 
 | |
5BSV
 
 | |
5BSM
 
 | |
5BSU
 
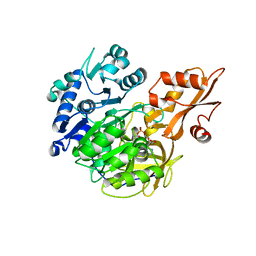 | | Crystal structure of 4-coumarate:CoA ligase complexed with caffeoyl adenylate | | Descriptor: | 4-coumarate--CoA ligase 2, 5'-O-[(R)-{[(2E)-3-(3,4-dioxocyclohexa-1,5-dien-1-yl)prop-2-enoyl]oxy}(hydroxy)phosphoryl]adenosine, GLYCEROL, ... | | Authors: | Li, Z, Nair, S.K. | | Deposit date: | 2015-06-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Specificity and Flexibility in a Plant 4-Coumarate:CoA Ligase.
Structure, 23, 2015
|
|
5BST
 
 | |
6L2C
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase in complex with CoA | | Descriptor: | Acetyl-CoA-acetyltransferase, putative, COENZYME A | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
1UAG
 
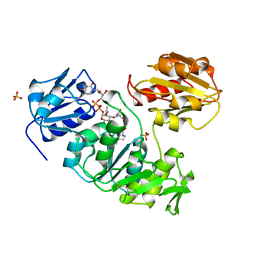 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE/:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J, Fanchon, E, Dideberg, O. | | Deposit date: | 1997-03-13 | | Release date: | 1998-03-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli.
EMBO J., 16, 1997
|
|
7PTH
 
 | |
7PTJ
 
 | |
5LID
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromopromazine | | Descriptor: | Cys-loop ligand-gated ion channel, bromopromazine | | Authors: | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | Deposit date: | 2016-07-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
