3ZLV
 
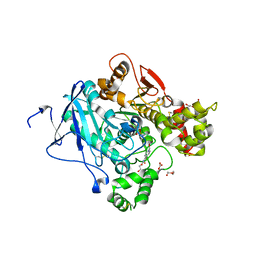 | | Crystal structure of mouse acetylcholinesterase in complex with tabun and HI-6 | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Artursson, E, Andersson, P.O, Akfur, C, Linusson, A, Borjegren, S, Ekstrom, F. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic-Site Conformational Equilibrium in Nerve-Agent Adducts of Acetylcholinesterase; Possible Implications for the Hi-6 Antidote Substrate Specificity.
Biochem.Pharmacol., 85, 2013
|
|
4QOC
 
 | |
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | Descriptor: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1HEH
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
5QED
 
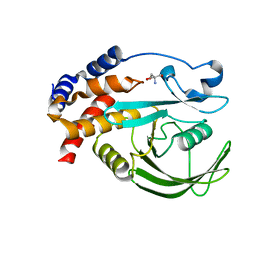 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000538a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-methyl-pyridine-2-carboxamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEV
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_XST00000603b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(4-chlorophenyl)-1H-pyrazol-5-amine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.723 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QFC
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000140a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-[(thiophen-2-yl)methyl]benzenesulfonamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
7X8R
 
 | | Cryo-EM structure of the Boc5-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-[(2-methylpropan-2-yl)oxycarbonylamino]phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7HH5
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z168883358 | | Descriptor: | 1,2-ETHANEDIOL, 1-(1,4-diazepan-1-yl)ethan-1-one, Hepatoma-derived growth factor-related protein 2, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
5QDI
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000157a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(2-phenylethyl)methanesulfonamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QDQ
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_XST00000847b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N,N-dimethylpyridin-4-amine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
6K5O
 
 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Kagechika, H, Ito, N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
3HV7
 
 | | Human p38 MAP Kinase in Complex with RL38 | | Descriptor: | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
1GVM
 
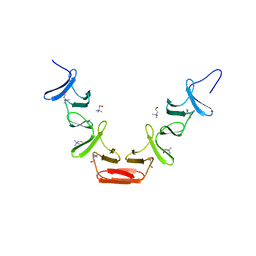 | | CHOLINE BINDING DOMAIN OF THE MAJOR AUTOLYSIN (C-LYTA) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AUTOLYSIN, CHOLINE ION, ... | | Authors: | Fernandez-Tornero, C, Lopez, R, Garcia, E, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2002-02-15 | | Release date: | 2002-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two New Crystal Forms of the Choline-Binding Domain of the Major Pneumococcal Autolysin: Insights Into the Dynamics of the Active Dimeric
J.Mol.Biol., 321, 2002
|
|
2WYT
 
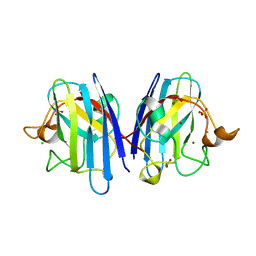 | | 1.0 A resolution structure of L38V SOD1 mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-20 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
6A2W
 
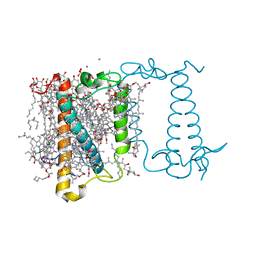 | | Crystal structure of fucoxanthin chlorophyll a/c complex from Phaeodactylum tricornutum | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W, Yu, L.J, Kuang, T.Y, Shen, J.R. | | Deposit date: | 2018-06-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for blue-green light harvesting and energy dissipation in diatoms.
Science, 363, 2019
|
|
2AYU
 
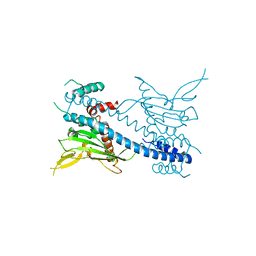 | |
9OR1
 
 | | N-hydroxylamine dehydratase (NohD) H2F/F4P/P5S/R6Y/A59N/R144Y/V96A/D172E/L172Q/D173P/R176V (P1/P2/A1/A2/A3) mutant crystal structure with heme and N-hydroxylated ornithine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-hydroxylamine dehydratase (NohD), N~5~-hydroxy-L-ornithine, ... | | Authors: | Higgins, M.A, Shi, X, Hoffarth, E.R, Du, Y.L, Ryan, K.S. | | Deposit date: | 2025-05-21 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conversion of a Heme-Dependent Dehydratase to a Piperazate Synthase Reveals the Role of the Heme Propionate Group in N-N Bond-Formation.
J.Am.Chem.Soc., 147, 2025
|
|
4YH7
 
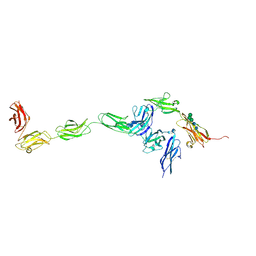 | | Crystal structure of PTPdelta ectodomain in complex with IL1RAPL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
3ZHF
 
 | | gamma 2 adaptin EAR domain crystal structure with preS1 site1 peptide NPDWDFN | | Descriptor: | 1,2-ETHANEDIOL, AP-1 COMPLEX SUBUNIT GAMMA-LIKE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Juergens, M.C, Voros, J, Rautureau, G, Shepherd, D, Pye, V.E, Muldoon, J, Johnson, C.M, Ashcroft, A, Freund, S.M.V, Ferguson, N. | | Deposit date: | 2012-12-21 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Hepatitis B Virus Pres1 Domain Hijacks Host Trafficking Proteins by Motif Mimicry.
Nat.Chem.Biol., 9, 2013
|
|
4IG9
 
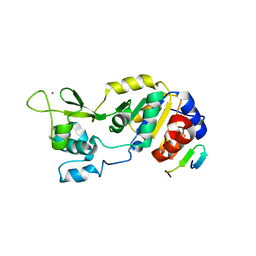 | |
1QQJ
 
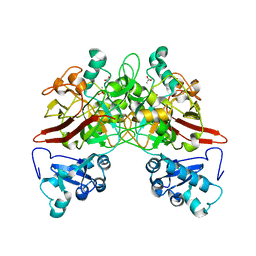 | | CRYSTAL STRUCTURE OF MOUSE FUMARYLACETOACETATE HYDROLASE REFINED AT 1.55 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
5QAU
 
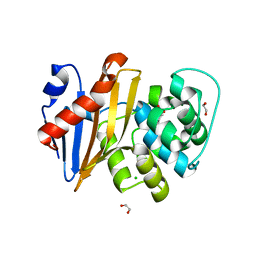 | | OXA-48 IN COMPLEX WITH COMPOUND 26a | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
1GYK
 
 | | Serum Amyloid P Component co-crystallised with MOBDG at neutral pH | | Descriptor: | CALCIUM ION, METHYL 4,6-O-[(1R)-1-CARBOXYETHYLIDENE]-BETA-D-GALACTOPYRANOSIDE, SERUM AMYLOID P-COMPONENT | | Authors: | Thompson, D, Pepys, M.B, Tickle, I, Wood, S.P. | | Deposit date: | 2002-04-25 | | Release date: | 2003-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structures of Crystalline Complexes of Human Serum Amyloid P Component with its Carbohydrate Ligand, the Cyclic Pyruvate Acetal of Galactose
J.Mol.Biol., 320, 2002
|
|
5QBP
 
 | | Crystal structure of Endothiapepsin-NAT17-347290a complex | | Descriptor: | 1-cyclohexyl-3-[(1S,2S,3S,4R,5R)-3-hydroxy-4-(piperidin-1-yl)-6,8-dioxabicyclo[3.2.1]octan-2-yl]urea, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Huschmann, F. | | Deposit date: | 2017-08-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Crystal structure of Endothiapepsin
To be published
|
|
