1K0O
 
 | | Crystal structure of a soluble form of CLIC1. An intracellular chloride ion channel | | Descriptor: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1 | | Authors: | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
2H62
 
 | | Crystal structure of a ternary ligand-receptor complex of BMP-2 | | Descriptor: | Acvr2b protein, Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Mueller, T.D. | | Deposit date: | 2006-05-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor.
Bmc Struct.Biol., 7, 2007
|
|
1JPS
 
 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
1JY1
 
 | | CRYSTAL STRUCTURE OF HUMAN TYROSYL-DNA PHOSPHODIESTERASE (TDP1) | | Descriptor: | TYROSYL-DNA PHOSPHODIESTERASE | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 2001-09-10 | | Release date: | 2002-02-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1.
Structure, 10, 2002
|
|
2H54
 
 | |
2H64
 
 | |
2P8C
 
 | | Crystal structure of N-succinyl Arg/Lys racemase from Bacillus cereus ATCC 14579 complexed with N-succinyl Arg. | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE | | Authors: | Fedorov, A.A, Song, L, Fedorov, E.V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction and assignment of function for a divergent N-succinyl amino acid racemase.
Nat.Chem.Biol., 3, 2007
|
|
5D4L
 
 | | Structure of the apo form of CPII from Thiomonas intermedia K12, a nitrogen regulatory PII-like protein | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
1LCL
 
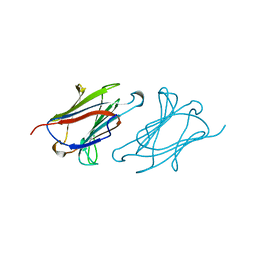 | | CHARCOT-LEYDEN CRYSTAL PROTEIN | | Descriptor: | LYSOPHOSPHOLIPASE | | Authors: | Acharya, K.R, Leonidas, D.D. | | Deposit date: | 1996-01-18 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Charcot-Leyden crystal protein, an eosinophil lysophospholipase, identifies it as a new member of the carbohydrate-binding family of galectins.
Structure, 3, 1995
|
|
2NDP
 
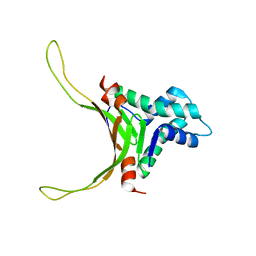 | | Structure of DNA-binding HU protein from micoplasma Mycoplasma gallisepticum | | Descriptor: | Histone-like DNA-binding superfamily protein | | Authors: | Altukhov, D.A, Talyzina, A.A, Agapova, Y.K, Vlaskina, A.V, Korzhenevskiy, D.A, Bocharov, E.V, Rakitina, T.V, Timofeev, V.I, Popov, V.O. | | Deposit date: | 2016-09-13 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enhanced conformational flexibility of the histone-like (HU) protein from Mycoplasma gallisepticum.
J.Biomol.Struct.Dyn., 36, 2018
|
|
2H4Y
 
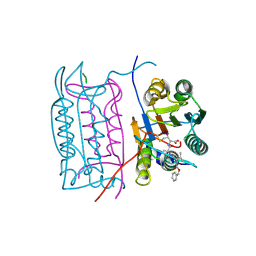 | |
2H51
 
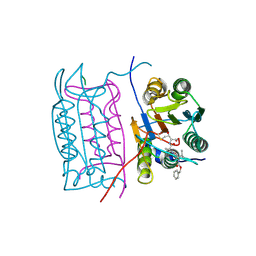 | |
4F42
 
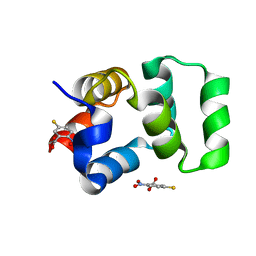 | | Neurotrophin p75NTR intracellular domain | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Qu, Q, Jiang, T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural insights into the allosteric activation mechanism of p75NTR
To be Published
|
|
3OSK
 
 | | Crystal structure of human CTLA-4 apo homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Yu, C, Sonnen, A.F.-P, Ikemizu, S, Stuart, D.I, Gilbert, R.J.C, Davis, S.J. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering
J.Biol.Chem., 286, 2011
|
|
4F44
 
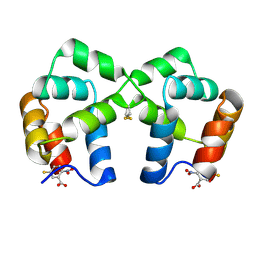 | | Neurotrophin p75NTR intracellular domain | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Qu, Q, Jiang, T. | | Deposit date: | 2012-05-10 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the allosteric activation mechanism of p75NTR
To be Published
|
|
2DEF
 
 | |
4F8T
 
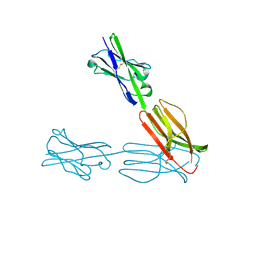 | | Crystal Structure of the Human BTN3A3 Ectodomain | | Descriptor: | Butyrophilin subfamily 3 member A3 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell response by CD277/Butryophilin (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4F9L
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 20.1 Single Chain Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 20.1 anti-BTN3A1 antibody fragment, Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1404 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
3GYU
 
 | | Nuclear receptor DAF-12 from parasitic nematode Strongyloides stercoralis in complex with its physiological ligand dafachronic acid delta 7 | | Descriptor: | (5beta,14beta,17alpha,25R)-3-oxocholest-7-en-26-oic acid, Nuclear hormone receptor of the steroid/thyroid hormone receptors superfamily, SRC1 | | Authors: | Zhou, X.E, Wang, Z, Suino-Powell, K, Motola, D.L, Conneely, A, Ogata, C, Sharma, K.K, Auchus, R.J, Kliewer, S.A, Xu, H.E, Mangelsdorf, D.J. | | Deposit date: | 2009-04-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | Descriptor: | GLUTAREDOXIN 3, GLUTATHIONE | | Authors: | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-30 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
2JJV
 
 | | Structure of human signal regulatory protein (sirp) beta(2) | | Descriptor: | CHLORIDE ION, SIGNAL-REGULATORY PROTEIN BETA 1., SULFATE ION | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Paired Receptor Specificity Explained by Structures of Signal Regulatory Proteins Alone and Complexed with Cd47.
Mol.Cell, 31, 2008
|
|
3GON
 
 | | Streptococcus pneumoniae Phosphomevalonate Kinase in Complex with Phosphomevalonate and AMPPNP | | Descriptor: | (3R)-3-HYDROXY-3-METHYL-5-(PHOSPHONOOXY)PENTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Andreassi, J.L, Bilder, P.W, Vetting, M.W, Roderick, S.L, Leyh, T.S. | | Deposit date: | 2009-03-19 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ternary complex of phosphomevalonate kinase: the enzyme and its family
Biochemistry, 48, 2009
|
|
2NQL
 
 | | Crystal structure of a member of the enolase superfamily from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, Isomerase/lactonizing enzyme, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Enolase from Agrobacterium Tumefaciens C58
To be Published
|
|
2E1Z
 
 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with diadenosine tetraphosphate (Ap4A) obtained after co-crystallization with ATP | | Descriptor: | 1,2-ETHANEDIOL, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Propionate Kinase | | Authors: | Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-11-04 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of Salmonella typhimurium propionate kinase and its complex with Ap4A: evidence for a novel Ap4A synthetic activity.
Proteins, 70, 2008
|
|
2E11
 
 | | The Crystal Structure of XC1258 from Xanthomonas campestris: A CN-hydrolase Superfamily Protein with an Arsenic Adduct in the Active Site | | Descriptor: | CACODYLATE ION, Hydrolase | | Authors: | Chin, K.-H, Tsai, Y.-D, Chan, N.-L, Huang, K.-F, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The crystal structure of XC1258 from Xanthomonas campestris: A putative procaryotic Nit protein with an arsenic adduct in the active site
Proteins, 69, 2007
|
|
