2O5Q
 
 | | Manganese horse heart myoglobin, nitric oxide modified | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, Myoglobin, NITRIC OXIDE, ... | | Authors: | Richter-Addo, G.B, Zahran, Z.N, Chooback, L, Copeland, D.M, West, A.H. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations.
J.Inorg.Biochem., 102, 2008
|
|
2O5S
 
 | | Cobalt horse heart myoglobin, nitrite modified | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Richter-Addo, G.B, Zahran, Z.N, Chooback, L, Copeland, D.M, West, A.H. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations.
J.Inorg.Biochem., 102, 2008
|
|
3SRN
 
 | | STRUCTURAL CHANGES THAT ACCOMPANY THE REDUCED CATALYTIC EFFICIENCY OF TWO SEMISYNTHETIC RIBONUCLEASE ANALOGS | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Martin, P.D, Doscher, M.S, Edwards, B.F.P. | | Deposit date: | 1991-05-20 | | Release date: | 1994-12-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes that accompany the reduced catalytic efficiency of two semisynthetic ribonuclease analogs.
J.Biol.Chem., 267, 1992
|
|
2YK0
 
 | | Structure of the N-terminal NTS-DBL1-alpha and CIDR-gamma double domain of the PfEMP1 protein from Plasmodium falciparum varO strain. | | Descriptor: | ERYTHROCYTE MEMBRANE PROTEIN 1, MAGNESIUM ION, SULFATE ION | | Authors: | Lewit-Bentley, A, Juillerat, A, Vigan-Womas, I, Guillotte, M, Hessel, A, Raynal, B, Mercereau-Puijalon, O, Bentley, G.A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Abo Blood-Group Dependence of Plasmodium Falciparum Rosetting.
Plos Pathog., 8, 2012
|
|
2P7C
 
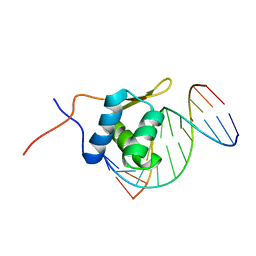 | | Solution structure of the bacillus licheniformis BlaI monomeric form in complex with the blaP half-operator. | | Descriptor: | Penicillinase repressor, Strand 1 of Twelve base-pair DNA, Strand 2 of Twelve base-pair DNA | | Authors: | Boudet, J, Duval, V, Van Melckebeke, H, Blackledge, M, Amoroso, A, Joris, B, Simorre, J.-P. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational and thermodynamic changes of the repressor/DNA operator complex upon monomerization shed new light on regulation mechanisms of bacterial resistance against beta-lactam antibiotics.
Nucleic Acids Res., 35, 2007
|
|
4GEY
 
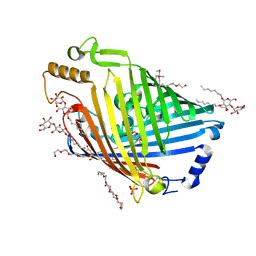 | | High pH structure of Pseudomonas putida OprB | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, DECYL-BETA-D-MALTOPYRANOSIDE, PHOSPHATE ION, ... | | Authors: | van den Berg, B. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for outer membrane sugar uptake in pseudomonads.
J.Biol.Chem., 287, 2012
|
|
1YLT
 
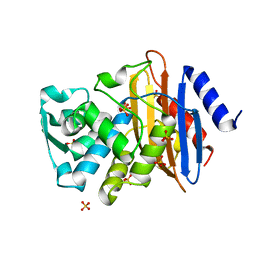 | | Atomic resolution structure of CTX-M-14 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-14 | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1YOL
 
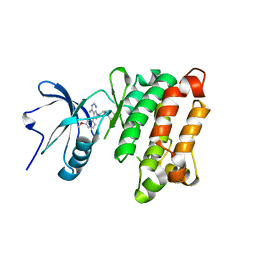 | | Crystal structure of Src kinase domain in complex with CGP77675 | | Descriptor: | 1-{4-[4-AMINO-5-(3-METHOXYPHENYL)-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL]BENZYL}PIPERIDIN-4-OL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | Deposit date: | 2005-01-27 | | Release date: | 2006-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
3SGS
 
 | |
1YOM
 
 | | Crystal structure of Src kinase domain in complex with Purvalanol A | | Descriptor: | 2-({6-[(3-CHLOROPHENYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)-3-METHYLBUTAN-1-OL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | Deposit date: | 2005-01-27 | | Release date: | 2006-01-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
3ZDL
 
 | | Vinculin head (1-258) in complex with a RIAM fragment | | Descriptor: | AMYLOID BETA A4 PRECURSOR PROTEIN-BINDING FAMILY B MEMBER 1-INTERACTING PROTEIN, VINCULIN | | Authors: | Zacharchenko, T, Elliott, P.R, Goult, B.T, Bate, N, Critchely, D.R, Barsukov, I.L. | | Deposit date: | 2012-11-28 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Riam and Vinculin Binding to Talin are Mutually Exclusive and Regulate Adhesion Assembly and Turnover.
J.Biol.Chem., 288, 2013
|
|
1XTO
 
 | | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6 | | Descriptor: | Coenzyme PQQ synthesis protein B, ZINC ION | | Authors: | Forouhar, F, Chen, Y, Kuzin, A, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-22 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6
To be Published
|
|
1MSG
 
 | |
2EPO
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | ACETIC ACID, N-acetyl-beta-D-glucosaminidase | | Authors: | Langley, D.B. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2OV6
 
 | |
2ZMM
 
 | | Crystal structure of PTP1B-inhibitor complex | | Descriptor: | 4-bromo-3-(carboxymethoxy)-5-{3-[cyclohexyl(methylcarbamoyl)amino]phenyl}thiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, W, Wu, J. | | Deposit date: | 2008-04-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Chemmedchem, 3, 2008
|
|
1N7X
 
 | | HUMAN SERUM TRANSFERRIN, N-LOBE Y45E MUTANT | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, Macgillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-18 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | THE POSITION OF ARGININE 124 CONTROLS THE RATE OF IRON RELEASE FROM THE N-LOBE OF HUMAN SERUM TRANSFERRIN. A STRUCTURAL STUDY
J.Biol.Chem., 278, 2003
|
|
3PTQ
 
 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 with dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, Beta-glucosidase Os4BGlu12, GLYCEROL, ... | | Authors: | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | Deposit date: | 2010-12-03 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
3Q2O
 
 | | Crystal Structure of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | MAGNESIUM ION, Phosphoribosylaminoimidazole carboxylase, ATPase subunit | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2010-12-20 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of N(5)-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1JBZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT HIGH PH | | Descriptor: | 1,2-ETHANEDIOL, GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
3UO0
 
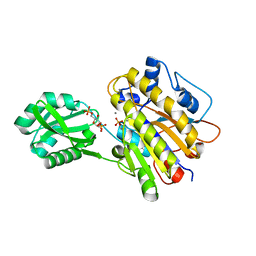 | | phosphorylated Bacillus cereus phosphopentomutase soaked with glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, ... | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3ZOR
 
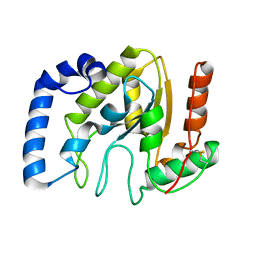 | | Structure of BsUDG | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Banos-Sanz, J.I, Mojardin, L, Sanz-Aparicio, J, Gonzalez, B, Salas, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure and Functional Insights Into Uracil-DNA Glycosylase Inhibition by Phage Phi29 DNA Mimic Protein P56
Nucleic Acids Res., 41, 2013
|
|
1KVQ
 
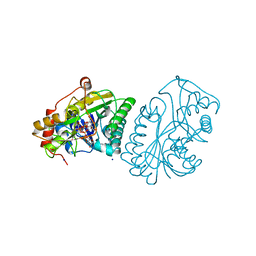 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1YBO
 
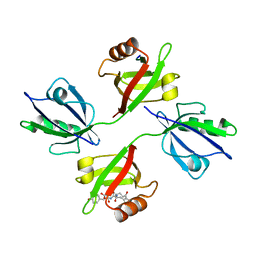 | | Crystal structure of the PDZ tandem of human syntenin with syndecan peptide | | Descriptor: | Syndecan-4, Syntenin 1 | | Authors: | Grembecka, J, Cooper, D.R, Cierpicki, T, Kang, B.S, Devedjiev, Y, Derewenda, Z. | | Deposit date: | 2004-12-21 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The binding of the PDZ tandem of syntenin to target proteins
Biochemistry, 45, 2006
|
|
3SGO
 
 | |
