1HSM
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
2GNX
 
 | | X-ray structure of a hypothetical protein from Mouse Mm.209172 | | Descriptor: | hypothetical protein | | Authors: | Phillips Jr, G.N, McCoy, J.G, Bitto, E, Wesenberg, G.E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-11 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray structure of a hypothetical protein from Mouse Mm.209172
To be Published
|
|
2H1S
 
 | |
3ULS
 
 | | Crystal structure of Fab12 | | Descriptor: | Fab12 heavy chain, Fab12 light chain | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3UF6
 
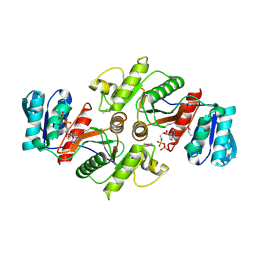 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A) | | Descriptor: | DEPHOSPHO COENZYME A, Lmo1369 protein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A)
To be Published
|
|
1IDX
 
 | |
2VJ0
 
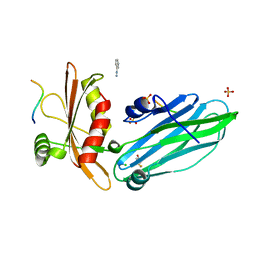 | | Crystal structure of the alpha-adaptin appendage domain, from the AP2 adaptor complex, in complex with an FXDNF peptide from amphiphysin1 and a WVXF peptide from synaptojanin P170 | | Descriptor: | AMPHIPHYSIN, AP-2 COMPLEX SUBUNIT ALPHA-2, BENZAMIDINE, ... | | Authors: | Ford, M.G.J, Praefcke, G.J.K, McMahon, H.T. | | Deposit date: | 2007-12-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Solitary and Repetitive Binding Motifs for the Ap2 Complex {Alpha}-Appendage in Amphiphysin and Other Accessory Proteins.
J.Biol.Chem., 283, 2008
|
|
2Z67
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) selenium transferase (SepSecS) | | Descriptor: | O-phosphoseryl-tRNA(Sec) selenium transferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Araiso, Y, Ishitani, R, Pailouer, S, Oshikane, H, Domae, N, Soll, D, Nureki, O. | | Deposit date: | 2007-07-23 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation.
Nucleic Acids Res., 36, 2008
|
|
2W3Z
 
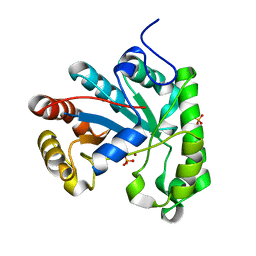 | | Structure of a Streptococcus mutans CE4 esterase | | Descriptor: | PHOSPHATE ION, PUTATIVE DEACETYLASE, ZINC ION | | Authors: | Deng, D.M, Urch, J.E, ten Cate, J.M, Rao, V.A, van Aalten, D.M.F, Crielaard, W. | | Deposit date: | 2008-11-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Streptococcus Mutans Smu.623C Codes for a Functional, Metal Dependent Polysaccharide Deacetylase that Modulates Interactions with Salivary Agglutinin.
J.Bacteriol., 191, 2009
|
|
2VYA
 
 | | Crystal Structure of fatty acid amide hydrolase conjugated with the drug-like inhibitor PF-750 | | Descriptor: | 4-(quinolin-3-ylmethyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Johnson, D.S, Wang, Z, Everdeen, D.S, Liimatta, M, Pabst, B, Bhattacharya, K, Nugent, R.A, Kamtekar, S, Cravatt, B.F, Ahn, K, Stevens, R.C. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Guided Inhibitor Design for Human Faah by Interspecies Active Site Conversion.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2W5O
 
 | | Complex structure of the GH93 alpha-L-arabinofuranosidase of Fusarium graminearum with arabinobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carapito, R, Imberty, A, Jeltsch, J.M, Byrns, S.C, Tam, P.H, Lowary, T.L, Varrot, A, Phalip, V. | | Deposit date: | 2008-12-11 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Basis of Arabinobio-Hydrolase Activity in Phytopathogenic Fungi. Crystal Structure and Catalytic Mechanism of Fusarium Graminearum Gh93 Exo-Alpha-L-Arabinanase.
J.Biol.Chem., 284, 2009
|
|
2WFV
 
 | | Crystal structure of DILP5 variant C4 | | Descriptor: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | Authors: | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | Deposit date: | 2009-04-15 | | Release date: | 2010-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
2X93
 
 | | Crystal structure of AnCE-trandolaprilat complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ANGIOTENSIN CONVERTING ENZYME, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
2X2E
 
 | | Dynamin GTPase dimer, long axis form | | Descriptor: | DYNAMIN-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chappie, J.S, Acharya, S, Leonard, M, Schmid, S.L, Dyda, F. | | Deposit date: | 2010-01-12 | | Release date: | 2010-04-28 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | G Domain Dimerization Controls Dynamin'S Assembly-Stimulated Gtpase Activity.
Nature, 465, 2010
|
|
3A1S
 
 | | Crystal structue of the cytosolic domain of T. maritima FeoB iron iransporter in GDP form I | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter
Structure, 17, 2009
|
|
3A1U
 
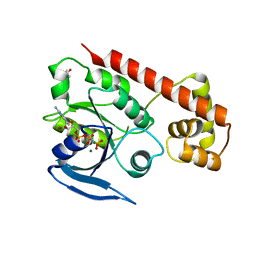 | | Crystal structue of the cytosolic domain of T. maritima FeoB iron iransporter in GMPPNP form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Iron(II) transport protein B, MAGNESIUM ION, ... | | Authors: | Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter
Structure, 17, 2009
|
|
2V8T
 
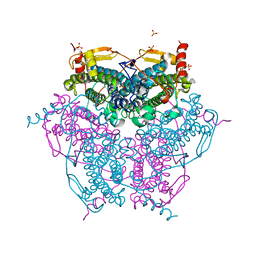 | | Crystal structure of Mn catalase from Thermus Thermophilus complexed with chloride | | Descriptor: | CHLORIDE ION, LITHIUM ION, MANGANESE (II) ION, ... | | Authors: | Antonyuk, S.V, Barynin, V.V, Vaguine, A.A, Melik-Adamyan, W.R, Popov, A.N, Lamsin, V.S, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Three-Dimentional Structure of the Enzyme Dimanganese Catalase from Thermus Thermophilus at 1 Angstrom Resolution
Crystallogr.Rep.(Transl. Kristallografiya), 45, 2000
|
|
3A2C
 
 | | Crystal structure of a pyrazolopyrimidine inhibitor complex bound to MAPKAP Kinase-2 (MK2) | | Descriptor: | MAP kinase-activated protein kinase 2, N~7~-(4-ethoxyphenyl)-6-methyl-N~5~-[(3S)-piperidin-3-yl]pyrazolo[1,5-a]pyrimidine-5,7-diamine, SULFATE ION | | Authors: | Fujino, A, Takimoto-Kamimura, M. | | Deposit date: | 2009-05-12 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of an MK2-inhibitor complex: insight into the regulation of the secondary structure of the Gly-rich loop by TEI-I01800
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1ZUN
 
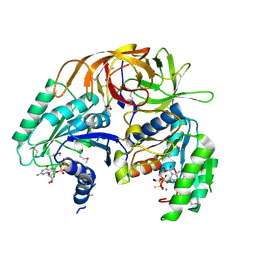 | | Crystal Structure of a GTP-Regulated ATP Sulfurylase Heterodimer from Pseudomonas syringae | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Mougous, J.D, Lee, D.H, Hubbard, S.C, Schelle, M.W, Vocadlo, D.J, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for g protein control of the prokaryotic ATP sulfurylase.
Mol.Cell, 21, 2006
|
|
3F38
 
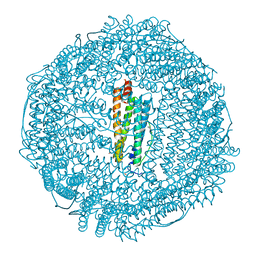 | | Apoferritin: complex with 2,6-dimethylphenol | | Descriptor: | 2,6-dimethylphenol, CADMIUM ION, Ferritin light chain, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
391D
 
 | | STRUCTURAL VARIABILITY AND NEW INTERMOLECULAR INTERACTIONS OF Z-DNA IN CRYSTALS OF D(PCPGPCPGPCPG) | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*G)-3') | | Authors: | Malinina, L, Tereshko, V, Ivanova, E, Subirana, J.A, Zarytova, V, Nekrasov, Y. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural variability and new intermolecular interactions of Z-DNA in crystals of d(pCpGpCpGpCpG).
Biophys.J., 74, 1998
|
|
2X2F
 
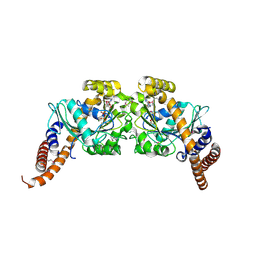 | | Dynamin 1 GTPase dimer, short axis form | | Descriptor: | DYNAMIN-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chappie, J.S, Acharya, S, Leonard, M, Schmid, S.L, Dyda, F. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | G Domain Dimerization Controls Dynamin'S Assembly-Stimulated Gtpase Activity.
Nature, 465, 2010
|
|
1O7Z
 
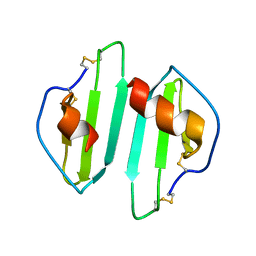 | |
2X0K
 
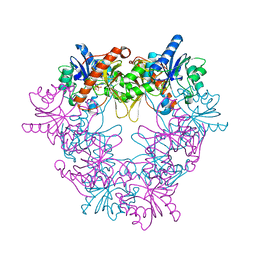 | | Crystal structure of modular FAD synthetase from Corynebacterium ammoniagenes | | Descriptor: | PYROPHOSPHATE, RIBOFLAVIN BIOSYNTHESIS PROTEIN RIBF, SULFATE ION | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Medina, M, Frago, S. | | Deposit date: | 2009-12-15 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligomeric State in the Crystal Structure of Modular Fad Synthetase Provides Insights Into its Sequential Catalysis in Prokaryotes
J.Mol.Biol., 400, 2010
|
|
3F32
 
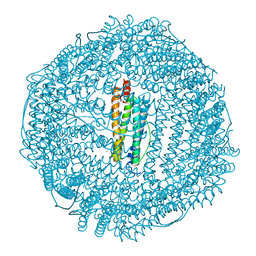 | | Horse spleen apoferritin | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
