1DPL
 
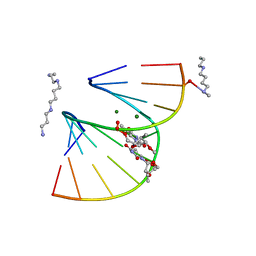 | | A-DNA DECAMER GCGTA(T23)TACGC WITH INCORPORATED 2'-METHOXY-3'-METHYLENEPHOSPHATE-THYMIDINE | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(T23)P*AP*CP*GP*C)-3', MAGNESIUM ION, SPERMINE | | Authors: | Egli, M, Tereshko, V, Teplova, M, Minasov, G, Joachimiak, A, Sanishvili, R, Weeks, C.M, Miller, R, Maier, M.A, An, H, Dan Cook, P, Manoharan, M. | | Deposit date: | 1999-12-27 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution.
Biopolymers, 48, 1998
|
|
6RAR
 
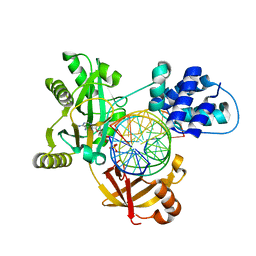 | | Pmar-Lig_PreS3-Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA (5'-D(*TP*TP*CP*CP*GP*AP*CP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Leiros, H.K.S, Williamson, A. | | Deposit date: | 2019-04-07 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
6XG8
 
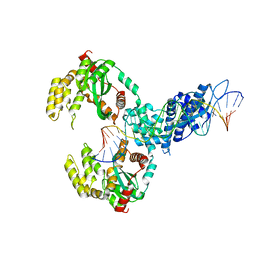 | | ISCth4 transposase, pre-cleaved complex, PCC | | Descriptor: | DNA (26-MER), Mutator family transposase | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
202D
 
 | |
5FFJ
 
 | |
6TC9
 
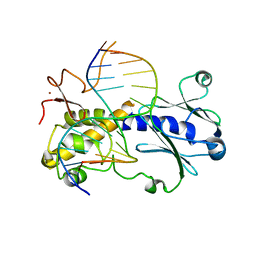 | | Crystal structure of MutM from Neisseria meningitidis | | Descriptor: | DNA, DNA containing abasic site analogue, Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Silhan, J, Landova, B, Boura, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Conformational changes of DNA repair glycosylase MutM triggered by DNA binding.
Febs Lett., 594, 2020
|
|
7XLT
 
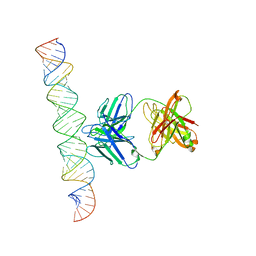 | | Cryo-EM Structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids | | Descriptor: | DNA, RNA, S9.6 Fab HC, ... | | Authors: | Li, Q, Lin, C, Luo, Z, Li, H, Li, X, Sun, Q. | | Deposit date: | 2022-04-22 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids.
J Genet Genomics, 49, 2022
|
|
3JR4
 
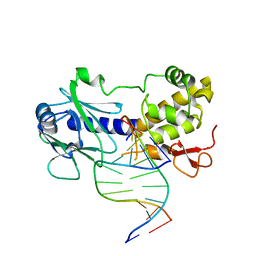 | | MutM interrogating an extrahelical G | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*G*CP*GP*TP*CP*CP*AP*(GX1)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Qi, Y, Spong, M.C, Verdine, G.L. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM.
J.Biol.Chem., 285, 2010
|
|
8JCD
 
 | |
8JBX
 
 | |
8GH8
 
 | | RuvA Holliday junction DNA complex | | Descriptor: | DNA (34-MER), Holliday junction branch migration complex subunit RuvA | | Authors: | Rish, A.D, Fu, T. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | RuvA Holliday junction DNA complex
To Be Published
|
|
6FL1
 
 | | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP(FOX)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B, Ober, M, Carell, T. | | Deposit date: | 2018-01-25 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA
To Be Published
|
|
4IM9
 
 | |
1AC7
 
 | | STRUCTURAL FEATURES OF THE DNA HAIRPIN D(ATCCTAGTTATAGGAT): THE FORMATION OF A G-A BASE PAIR IN THE LOOP, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*TP*AP*GP*TP*TP*AP*TP*AP*GP*GP*AP*T)-3') | | Authors: | Van Dongen, M.J.P, Mooren, M.M.W, Willems, E.F.A, Van Der Marel, G.A, Van Boom, J.H, Wijmenga, S.S, Hilbers, C.W. | | Deposit date: | 1997-02-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of the DNA hairpin d(ATCCTA-GTTA-TAGGAT): formation of a G-A base pair in the loop.
Nucleic Acids Res., 25, 1997
|
|
4FS1
 
 | | Base pairing mechanism of N2,3-ethenoguanine with dTTP by human polymerase iota | | Descriptor: | DNA 5'-D(*TP*CP*TP*(EFG)P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*(DOC))-3', DNA polymerase iota, MAGNESIUM ION, ... | | Authors: | Zhao, L. | | Deposit date: | 2012-06-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Basis of Miscoding of the DNA Adduct N2,3-Ethenoguanine by Human Y-family DNA Polymerases.
J.Biol.Chem., 287, 2012
|
|
5EG0
 
 | | HOXB13-MEIS1 heterodimer bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*GP*TP*AP*AP*AP*AP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*G)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Molecular basis of recognition of two distinct DNA sequences by a single transcription factor
To Be Published
|
|
6SXA
 
 | | XPF-ERCC1 Cryo-EM Structure, Apo-form | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
1BAE
 
 | |
1VT8
 
 | | Crystal structure of D(GGGCGCCC)-hexagonal form | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | Deposit date: | 1996-12-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
6TC6
 
 | |
343D
 
 | |
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
7SWY
 
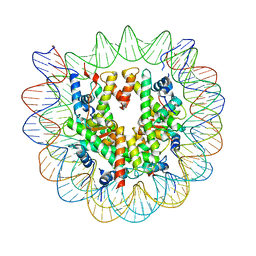 | | 2.6 A structure of a 40-601[TA-rich+1]-40 nucleosome | | Descriptor: | DNA Guide Strand, DNA Tracking Strand, Histone H2A, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1ZNA
 
 | |
2IUT
 
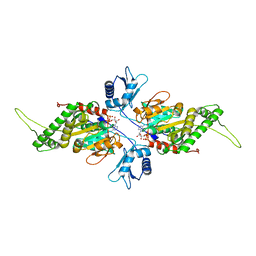 | | P. aeruginosa FtsK motor domain, dimeric | | Descriptor: | DNA TRANSLOCASE FTSK, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Massey, T.H, Mercogliano, C.P, Yates, J, Sherratt, D.J, Lowe, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric Ftsk
Mol.Cell, 23, 2006
|
|
