7QL5
 
 | | Torpedo muscle-type nicotinic acetylcholine receptor - nicotine-bound conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zarkadas, E, Pebay-Peyroula, E, Baenziger, J, Nury, H. | | Deposit date: | 2021-12-19 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Conformational transitions and ligand-binding to a muscle-type nicotinic acetylcholine receptor.
Neuron, 110, 2022
|
|
7QKO
 
 | | Torpedo muscle-type nicotinic acetylcholine receptor - Resting conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Acetylcholine receptor subunit alpha, Acetylcholine receptor subunit beta, ... | | Authors: | Zarkadas, E, Pebay-Peyroula, E, Baenziger, J, Nury, H. | | Deposit date: | 2021-12-18 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational transitions and ligand-binding to a muscle-type nicotinic acetylcholine receptor.
Neuron, 110, 2022
|
|
6MWZ
 
 | | LasR LBD T75V/Y93F/A127W:BB0126 | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1S,3S,5S)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, ALA-HIS-HIS-HIS-HIS-ALA, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
7QL6
 
 | | Torpedo muscle-type nicotinic acetylcholine receptor - carbamylcholine-bound conformation | | Descriptor: | 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zarkadas, E, Pebay-Peyroula, E, Baenziger, J, Nury, H. | | Deposit date: | 2021-12-19 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conformational transitions and ligand-binding to a muscle-type nicotinic acetylcholine receptor.
Neuron, 110, 2022
|
|
5IIY
 
 | | SrpA adhesin in complex with the Neu5Ac-galactoside disaccharide | | Descriptor: | ACETATE ION, CALCIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2016-03-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structures of the Streptococcus sanguinis SrpA Binding Region with Human Sialoglycans Suggest Features of the Physiological Ligand.
Biochemistry, 2016
|
|
5O75
 
 | | Ube4B U-box domain | | Descriptor: | SULFATE ION, Ubiquitin conjugation factor E4 B | | Authors: | Gabrielsen, M, Buetow, L, Nakasone, M.A, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
4ABQ
 
 | | CRYSTAL STRUCTURE OF TRANSTHYRETIN IN COMPLEX WITH LIGAND C-1 | | Descriptor: | 3-(5-MERCAPTO-1,3,4-OXADIAZOL-2-YL)-PHENOL, TRANSTHYRETIN | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-10 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
4ABV
 
 | | CRYSTAL STRUCTURE OF TRANSTHYRETIN IN COMPLEX WITH LIGAND C-3 | | Descriptor: | 5-(chloromethyl)-2-(2,4-dichlorophenoxy)phenol, TRANSTHYRETIN | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
4ABW
 
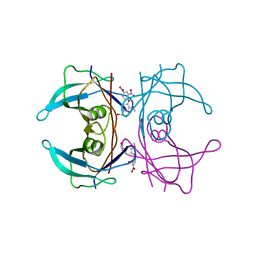 | | Crystal Structure of Transthyretin in Complex With Ligand C-6 | | Descriptor: | TRANSTHYRETIN, {4-[4-(hydroxymethyl)-2-methoxyphenoxy]benzene-1,3-diyl}bis[hydroxy(oxo)ammonium] | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
4ABU
 
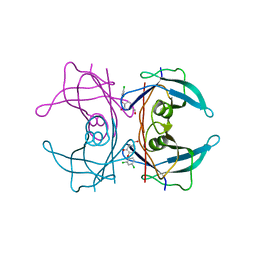 | | CRYSTAL STRUCTURE OF TRANSTHYRETIN IN COMPLEX WITH LIGAND C-2 | | Descriptor: | 4-(2,4-dichlorophenoxy)-3-hydroxybenzaldehyde, TRANSTHYRETIN | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
8EI3
 
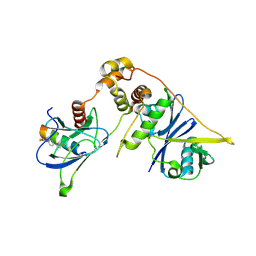 | | Crystal structure of VHL in complex with H313, a Helicon Polypeptide | | Descriptor: | Elongin-B, Elongin-C, H313, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
5KIQ
 
 | | SrpA with sialyl LewisX | | Descriptor: | ACETATE ION, CALCIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | Structures of the Streptococcus sanguinis SrpA Binding Region with Human Sialoglycans Suggest Features of the Physiological Ligand.
Biochemistry, 2016
|
|
5IJ3
 
 | | SrpA adhesin in complex with sialyl T antigen | | Descriptor: | ACETATE ION, CALCIUM ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2016-03-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the Streptococcus sanguinis SrpA Binding Region with Human Sialoglycans Suggest Features of the Physiological Ligand.
Biochemistry, 2016
|
|
1JOJ
 
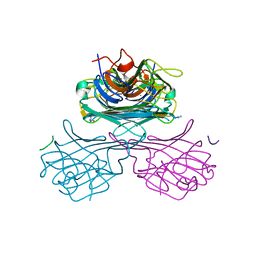 | | CONCANAVALIN A-HEXAPEPTIDE COMPLEX | | Descriptor: | CALCIUM ION, Concanavalin-Br, HEXAPEPTIDE, ... | | Authors: | Jain, D, Kaur, K, Salunke, D.M. | | Deposit date: | 2001-07-30 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhanced binding of a rationally designed peptide ligand of concanavalin a arises from improved geometrical complementarity.
Biochemistry, 40, 2001
|
|
1JZK
 
 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F mutant (deoxy) | | Descriptor: | GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
6T44
 
 | |
6Z4Q
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule inverse agonist SR142948A | | Descriptor: | 2-[[5-(2,6-dimethoxyphenyl)-1-[4-[3-(dimethylamino)propyl-methyl-carbamoyl]-2-propan-2-yl-phenyl]pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z8N
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule full agonist SRI-9829 | | Descriptor: | (2~{S})-4-methyl-2-[(1-quinolin-8-ylsulfonylindol-3-yl)carbonylamino]pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6YVR
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the peptide full agonist NTS8-13 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin crystallisation chaperone, neurotensin NTS8-13 (full agonist), nonyl beta-D-glucopyranoside | | Authors: | Deluigi, M, Merklinger, L, Hilge, M, Ernst, P, Klipp, A, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZA8
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule partial agonist RTI-3a | | Descriptor: | (2~{S})-2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]-4-methyl-pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1),Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4V
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with NTS8-13 | | Descriptor: | ARG-PRO-TYR-ILE-LEU, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
4FFE
 
 | | The structure of cowpox virus CPXV018 (OMCP) | | Descriptor: | CPXV018 protein | | Authors: | Lazear, E, Peterson, L.W, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Cowpox Virus-Encoded NKG2D Ligand OMCP.
J.Virol., 87, 2013
|
|
6T42
 
 | |
2ZHQ
 
 | | Thrombin Inhibition | | Descriptor: | Hirudin variant-1, N-(4-carbamimidoylbenzyl)-1-(3-phenylpropanoyl)-L-prolinamide, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-02-08 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Non-additivity of functional group contributions in protein-ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry.
J.Mol.Biol., 397, 2010
|
|
2JMO
 
 | | IBR domain of Human Parkin | | Descriptor: | Parkin, ZINC ION | | Authors: | Beasley, S.A, Hristova, V.A, Shaw, G.S. | | Deposit date: | 2006-11-24 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the Parkin in-between-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson's disease.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
