1VPB
 
 | |
2PLH
 
 | | STRUCTURE OF ALPHA-1-PUROTHIONIN AT ROOM TEMPERATURE AND 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2-BUTANOL, ACETATE ION, ALPHA-1-PUROTHIONIN, ... | | Authors: | Teeter, M.M, Stec, B, Rao, U. | | Deposit date: | 1993-07-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of purothionins reveals solute particles important for lattice formation and toxicity. Part 1: alpha1-purothionin revisited.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2PPL
 
 | | Human Pancreatic lipase-related protein 1 | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 1, SODIUM ION | | Authors: | Walker, J.R, Davis, T, Seitova, A, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-30 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Human Pancreatic Lipase-related Protein 1.
To be Published
|
|
5EAM
 
 | | Crystal structure of human WDR5 in complex with compound 9o | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
3CPX
 
 | |
3ATG
 
 | | endo-1,3-beta-glucanase from Cellulosimicrobium cellulans | | Descriptor: | CALCIUM ION, GLUCANASE, GLYCEROL, ... | | Authors: | Tanabe, Y, Pang, Z, Oda, M, Mikami, B. | | Deposit date: | 2011-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and thermodynamic characterization of endo-1,3-beta-glucanase: Insights into the substrate recognition mechanism.
Biochim. Biophys. Acta, 1866, 2018
|
|
1ZZM
 
 | | Crystal structure of YJJV, TATD Homolog from Escherichia coli k12, at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ZINC ION, putative deoxyribonuclease yjjV | | Authors: | Malashkevich, V.N, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-06-14 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YJJV, TATD homolog from Escherichia coli K12, at 1.8 A resolution
To be Published
|
|
4NTL
 
 | |
4NTO
 
 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C2 ceramide-1-phosphate (d18:1/2:0) at 2.15 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
1LV1
 
 | | Crystal Structure Analysis of the non-active site mutant of tethered HIV-1 protease to 2.1A resolution | | Descriptor: | HIV-1 protease | | Authors: | Kumar, M, Kannan, K.K, Hosur, M.V, Bhavesh, N.S, Chatterjee, A, Mittal, R, Hosur, R.V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
5ZGU
 
 | |
4Q7W
 
 | |
1LBL
 
 | | Crystal structure of indole-3-glycerol phosphate synthase (IGPS) in complex with 1-(o-carboxyphenylamino)-1-deoxyribulose 5'-phosphate (CdRP) | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, indole-3-glycerol phosphate synthase | | Authors: | Hennig, M, Darimont, B.D, Kirschner, K, Jansonius, J.N. | | Deposit date: | 2002-04-04 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The catalytic mechanism of indole-3-glycerol phosphate synthase: Crystal structures of complexes of the enzyme from Sulfolobus solfataricus with the substrate analogue, substrate, and product
J.Mol.Biol., 319, 2002
|
|
3K5G
 
 | | Human bace-1 complex with bjc060 | | Descriptor: | (1R,3S)-N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(1-methylethyl)benzyl]amino}propyl]-3-[1-methyl-1-(2-oxopiperidin-1-yl)ethy l]cyclohexanecarboxamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5ZKZ
 
 | |
3BQM
 
 | | LFA-1 I domain bound to inhibitors | | Descriptor: | 3-({4-[(1E)-3-morpholin-4-yl-3-oxoprop-1-en-1-yl]-2,3-bis(trifluoromethyl)phenyl}sulfanyl)aniline, Integrin alpha-L | | Authors: | Silvian, L.F. | | Deposit date: | 2007-12-20 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and synthesis of a series of meta aniline-based LFA-1 ICAM inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5ZFI
 
 | | Mouse kallikrein 7 in complex with 6-benzyl-1,4-diazepan-7-one derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[(3Z,6R)-6-[(2,6-dichlorophenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxo-1,4-diazepan-1-yl]-N-[3-(1-methyl-1H-pyrazol-4-yl)phenyl]acetamide, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
5UNJ
 
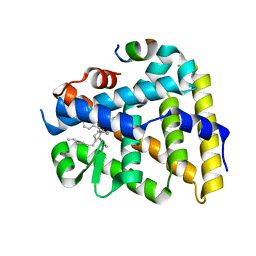 | | Structure of Human Liver Receptor Homolog 1 in complex with PGC1a and RJW100 | | Descriptor: | (1R,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor subfamily 5 group A member 2, Peroxisome proliferator-activated gamma coactivator 1-alpha | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Dynamics of the Liver Receptor Homolog 1-PGC1 alpha Complex.
Mol. Pharmacol., 92, 2017
|
|
3S25
 
 | |
3B9T
 
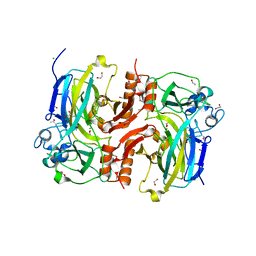 | |
1Q1C
 
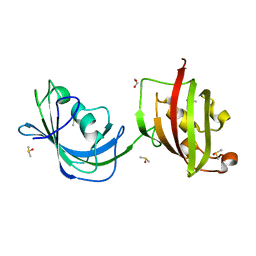 | | Crystal structure of N(1-260) of human FKBP52 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-07-18 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3C5Y
 
 | |
4N3A
 
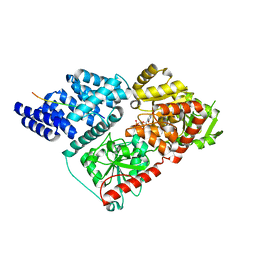 | |
2WWM
 
 | | Crystal structure of the Titin M10-Obscurin like 1 Ig complex in space group P1 | | Descriptor: | OBSCURIN-LIKE PROTEIN 1, TITIN | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight Into M-Band Assembly and Mechanics from the Titin-Obscurin-Like-1 Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3BVA
 
 | | Cystal structure of HIV-1 Active Site Mutant D25N and p2-NC analog inhibitor | | Descriptor: | GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, Protease (Retropepsin) | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
