6LPI
 
 | | Crystal Structure of AHAS holo-enzyme | | Descriptor: | Acetolactate synthase isozyme 1 large subunit, Acetolactate synthase isozyme 1 small subunit, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, Y, Yang, X, Xi, Z, Shen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Molecular architecture of the acetohydroxyacid synthase holoenzyme.
Biochem.J., 477, 2020
|
|
5YPP
 
 | | Crystal structure of IlvN.Val-1a | | Descriptor: | ACETATE ION, Acetolactate synthase isozyme 1 small subunit, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
5YPW
 
 | | Crystal structure of IlvN.Val-1b | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, VALINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-03 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
5YPY
 
 | | Crystal structure of IlvN. Val-1c | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, VALINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-04 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
4LVC
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine | | Descriptor: | ACETATE ION, ADENOSINE, AMMONIUM ION, ... | | Authors: | Manszewski, T, Singh, K, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An enzyme captured in two conformational states: crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6OHD
 
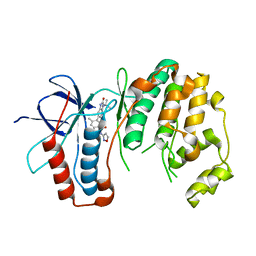 | | P38 in complex with T-3220137 | | Descriptor: | 3-(3-tert-butyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)-4-methyl-N-(1,2-oxazol-3-yl)benzamide, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]Pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 2.
Chemmedchem, 14, 2019
|
|
4WHU
 
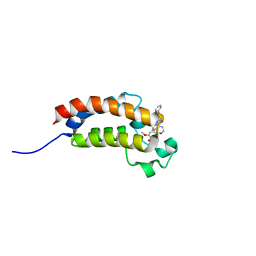 | | BROMO domain of CREB binding protein | | Descriptor: | 2-methoxy-4-{1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazol-5-yl}cyclohepta-2,4,6-trien-1-one, CREB-binding protein | | Authors: | Liu, S. | | Deposit date: | 2014-09-23 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Direct photocapture of bromodomains using tropolone chemical probes
To Be Published
|
|
2QWE
 
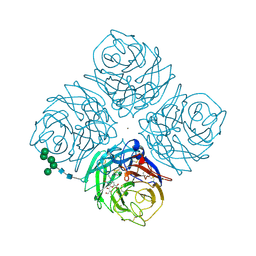 | |
2QWC
 
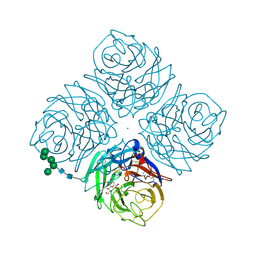 | |
2QWB
 
 | |
2QWD
 
 | |
5WLY
 
 | | E. coli LpxH- 8 mutations | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
6GHZ
 
 | |
6GI3
 
 | |
6GHY
 
 | |
3S0N
 
 | | Crystal Structure of Human Chymase with Benzimidazolone Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{3-[(4-methyl-1-benzothiophen-3-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}butanoic acid, Chymase, ... | | Authors: | Qian, K.C, Farrow, N.A, Padyana, A.K. | | Deposit date: | 2011-05-13 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Benzimidazolone as potent chymase inhibitor: Modulation of reactive metabolite formation in the hydrophobic (P(1)) region.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4K60
 
 | | Crystal Structure of Human Chymase in Complex with Fragment 6-bromo-1,3-dihydro-2H-indol-2-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromo-1,3-dihydro-2H-indol-2-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4K5Z
 
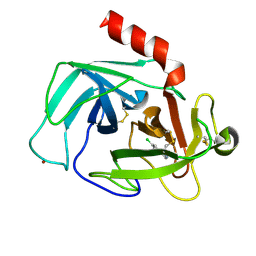 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-2,3-dihydro-1H-isoindol-1-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-2,3-dihydro-1H-isoindol-1-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4K69
 
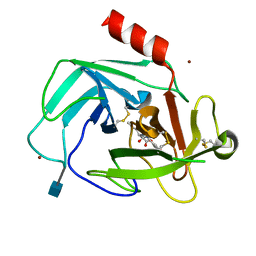 | | Crystal Structure of Human Chymase in Complex with Fragment Linked Benzimidazolone Inhibitor: (3S)-3-{3-[(6-bromo-2-oxo-2,3-dihydro-1H-indol-4-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}hexanoic acid | | Descriptor: | (3S)-3-{3-[(6-bromo-2-oxo-2,3-dihydro-1H-indol-4-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}hexanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
4UDB
 
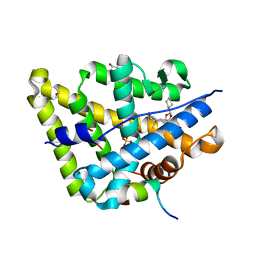 | | MR in complex with desisobutyrylciclesonide | | Descriptor: | DESISOBUYTYRYL CICLESONIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, JellesmarkJensen, T, Cavallin, A, Nilsson, E, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
6VHF
 
 | | Crystal structure of RbBP5 interacting domain of Cfp1 | | Descriptor: | PHD-type domain-containing protein, ZINC ION | | Authors: | Joshi, M, Couture, J.F. | | Deposit date: | 2020-01-09 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | A non-canonical monovalent zinc finger stabilizes the integration of Cfp1 into the H3K4 methyltransferase complex COMPASS.
Nucleic Acids Res., 48, 2020
|
|
6VME
 
 | | Human ESCRT-I heterotetramer headpiece | | Descriptor: | Multivesicular body subunit 12A, Tumor susceptibility gene 101 protein, Vacuolar protein sorting-associated protein 28 homolog, ... | | Authors: | Flower, T.G, Hurley, J.H, Tjahjono, N. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
7XMC
 
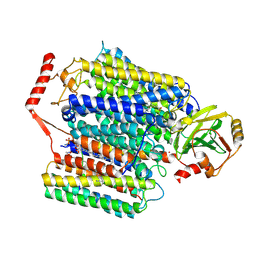 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, apo structure with DMSO | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMD
 
 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, the structure complexed with an allosteric inhibitor N4 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
