4WNJ
 
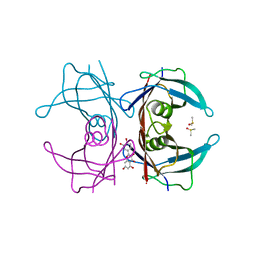 | | Crystal structure of Transthyretin-quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Berni, R, Folli, C. | | Deposit date: | 2014-10-13 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3FIR
 
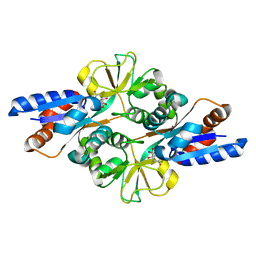 | | Crystal structure of Glycosylated K135E PEB3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-12 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
4WNS
 
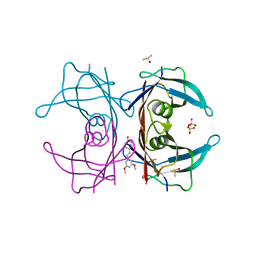 | | Crystal structure of Transthyretin complexed with pterostilbene | | Descriptor: | DIMETHYL SULFOXIDE, Pterostilbene, SULFATE ION, ... | | Authors: | Zanotti, G, Cianci, M, Folli, C, Berni, R. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7OHD
 
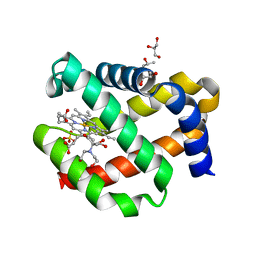 | | CRYSTAL STRUCTURE OF FERRIC MURINE NEUROGLOBIN CDLESS MUTANT | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Cerutti, G, Gugole, E, Vallone, B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Role of Murine Neuroglobin CDloop-D-Helix Unit in CO Ligand Binding and Structural Dynamics.
Acs Chem.Biol., 17, 2022
|
|
4RPJ
 
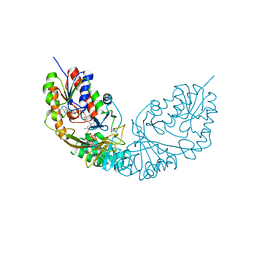 | |
7UTC
 
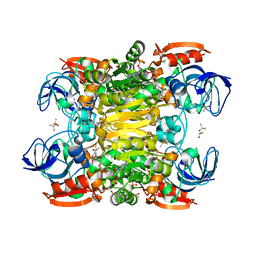 | |
7UUT
 
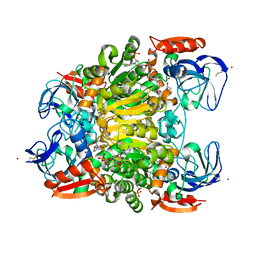 | | Ternary complex crystal structure of secondary alcohol dehydrogenases from the Thermoanaerobacter ethanolicus mutants C295A and I86A provides better understanding of catalytic mechanism | | Descriptor: | (2R)-pentan-2-ol, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Dinh, T, Phillips, R, Rahn, K. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic snapshots of ternary complexes of thermophilic secondary alcohol dehydrogenase from Thermoanaerobacter pseudoethanolicus reveal the dynamics of ligand exchange and the proton relay network.
Proteins, 90, 2022
|
|
6W4G
 
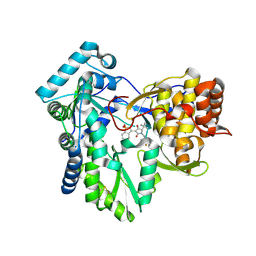 | | Hepatitis C virus polymerase NS5B with RO inhibitor for SAR studies | | Descriptor: | 5-[[(2~{S})-4-[1,1-bis(oxidanylidene)-1,2-benzothiazol-3-yl]-2-~{tert}-butyl-3-oxidanyl-5-oxidanylidene-2~{H}-pyrrol-1-yl]methyl]-2-fluoranyl-benzenecarbonitrile, NS5B | | Authors: | Harris, S.F. | | Deposit date: | 2020-03-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of Noncompetitive Protein-Ligand Interactions for Structural Optimization.
J.Chem.Inf.Model., 60, 2020
|
|
8DYC
 
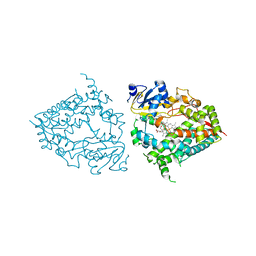 | | Human CYP3A4 bound to a substrate | | Descriptor: | 2-butyl-6-(butylamino)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-08-04 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of CYP3A4 Complexed with Fluorol Identifies the Substrate Access Channel as a High-Affinity Ligand Binding Site.
Int J Mol Sci, 23, 2022
|
|
1R1U
 
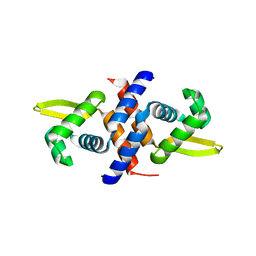 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the apo-form | | Descriptor: | repressor protein | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
6XZ1
 
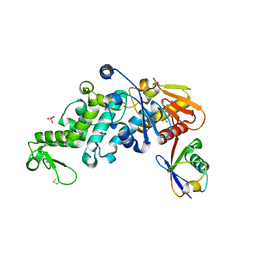 | | Conjugate of the HECT domain of HUWE1 with ubiquitin | | Descriptor: | HECT, UBA and WWE domain containing 1, isoform CRA_a, ... | | Authors: | Liu, B, Seenivasan, A, Nair, R, Chen, D, Lowe, E.D, Lorenz, S. | | Deposit date: | 2020-01-31 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution and Structural Analysis of a HECT Ligase-Ubiquitin Complex via an Activity-Based Probe.
Acs Chem.Biol., 16, 2021
|
|
4B5D
 
 | | Capitella teleta AChBP in complex with psychonicline (3-((2(S)- Azetidinyl)methoxy)-5-((1S,2R)-2-(2-hydroxyethyl)cyclopropyl)pyridine) | | Descriptor: | 2-[(1R,2S)-2-[5-[[(2S)-azetidin-2-yl]methoxy]pyridin-3-yl]cyclopropyl]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nys, M, Ulens, C. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Insights Into the Structural Determinants Required for High- Affinity Binding of Chiral Cyclopropane-Containing Ligands to Alpha4Beta2-Nicotinic Acetylcholine Receptors: An Integrated Approach to Behaviorally Active Nicotinic Ligands.
J.Med.Chem., 55, 2012
|
|
2P7A
 
 | |
8TQD
 
 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
8JDW
 
 | |
7ULD
 
 | |
7ULJ
 
 | | Hsp90b N-terminal domain in complex with 42C | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Heat shock protein HSP 90-beta, ... | | Authors: | Stachowski, T.R, Nithianantham, S, Vanarotti, M, Fischer, M. | | Deposit date: | 2022-04-05 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pan-HSP90 ligand binding reveals isoform-specific differences in plasticity and water networks.
Protein Sci., 32, 2023
|
|
7ULF
 
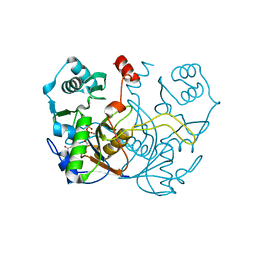 | |
2J9X
 
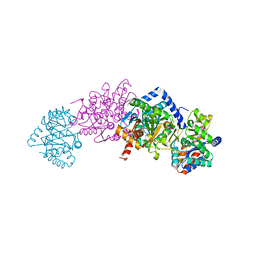 | | Tryptophan Synthase in complex with GP, alpha-D,L-glycerol-phosphate, Cs, pH6.5 - alpha aminoacrylate form - (GP)E(A-A) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ngo, H, Kimmich, N, Harris, R, Niks, D, Blumenstein, L, Kulik, V, Barends, T.R, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-11-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Regulation of Substrate Channeling in Tryptophan Synthase: Modulation of the L-Serine Reaction in Stage I of the Beta-Reaction by Alpha-Site Ligands.
Biochemistry, 46, 2007
|
|
5LI8
 
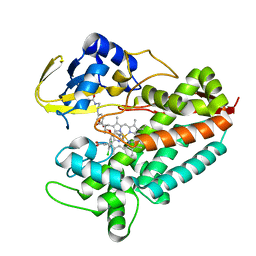 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with ketoconazole | | Descriptor: | 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126 | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
4KFQ
 
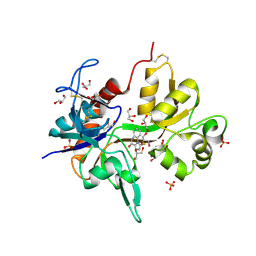 | | Crystal structure of the NMDA receptor GluN1 ligand binding domain in complex with 1-thioxo-1,2-dihydro-[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one | | Descriptor: | 1-sulfanyl[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Steffensen, T.B, Tabrizi, F.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site.
J.Biol.Chem., 288, 2013
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
5LIE
 
 | |
5LI7
 
 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with 1-(3-(1H-imidazol-1-yl)propyl)-3-((3s,5s,7s)-adamantan-1-yl)urea | | Descriptor: | 1-(1-adamantyl)-3-(3-imidazol-1-ylpropyl)urea, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126 | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
3GUP
 
 | |
