2K69
 
 | |
2K68
 
 | |
2K67
 
 | |
2K62
 
 | | NMR solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based Gd(III)-chelate | | Descriptor: | (3alpha,5alpha,8alpha)-3-[(N,N-bis{2-[bis(carboxymethyl)amino]ethyl}-L-gamma-glutamyl)amino]cholan-24-oic acid, Liver fatty acid-binding protein, YTTERBIUM (III) ION | | Authors: | Tomaselli, S, Zanzoni, S, Ragona, L, Gianolio, E, Aime, S, Assfalg, M, Molinari, H. | | Deposit date: | 2008-07-03 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based gadolinium(III)-chelate, a potential hepatospecific magnetic resonance imaging contrast agent.
J.Med.Chem., 51, 2008
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2KFY
 
 | |
2KVY
 
 | | NMR solution structure of the 4:1 complex between an uncharged distamycin A analogue and [d(TGGGGT)]4 | | Descriptor: | 4-amino-1-methyl-N-{1-methyl-5-[(1-methyl-5-{[3-(methylamino)-3-oxopropyl]carbamoyl}-1H-pyrrol-3-yl)carbamoyl]-1H-pyrrol-3-yl}-1H-pyrrole-2-carboxamide, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3') | | Authors: | Cosconati, S, Marinelli, L, Trotta, R, Virno, A, De Tito, S, Romagnoli, R, Pagano, B, Limongelli, V, Giancola, C, Baraldi, P, Mayol, L, Novellino, E, Randazzo, A. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle.
J.Am.Chem.Soc., 132, 2010
|
|
2KHK
 
 | | NMR solution structure of the b30-82 domain of subunit b of Escherichia coli F1FO ATP synthase | | Descriptor: | ATP synthase subunit b | | Authors: | Priya, R, Biukovic, G, Gayen, S, Vivekanandan, S, Gruber, G. | | Deposit date: | 2009-04-08 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure, determined by nuclear magnetic resonance, of the b30-82 domain of subunit b of Escherichia coli F1Fo ATP synthase
J.Bacteriol., 191, 2009
|
|
2L0K
 
 | | NMR solution structure of a transcription factor SpoIIID in complex with DNA | | Descriptor: | Stage III sporulation protein D | | Authors: | Chen, B, Himes, P, Lu, Z, Liu, A, Yan, H, Kroos, L. | | Deposit date: | 2010-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Novel Mode of DNA Binding by Bacterial Transcription Factor SpoIIID
To be Published
|
|
2L1Y
 
 | | NMR Structure of human insulin mutant GLY-B20-D-ALA, GLY-B23-D-ALA PRO-B28-LYS, LYS-B29-PRO, 20 Structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wan, Z.L, Hua, Q.X, Huang, K, Hu, S.Q, Philips, N.B, Katsoyannis, J.W, Weiss, M.A. | | Deposit date: | 2010-08-09 | | Release date: | 2011-08-31 | | Method: | SOLUTION NMR | | Cite: | Chiral Protein Engineering and its Application in G Health
To be Published
|
|
2L1Z
 
 | | NMR Structure of human insulin mutant GLY-B20-D-ALA, GLY-B23-D-ALA PRO-B28-LYS, LYS-B29-PRO, 20 Structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wan, Z.L, Hua, Q.X, Huang, K, Hu, S.Q, Philips, N.B, Katsoyannis, J.W, Weiss, M.A. | | Deposit date: | 2010-08-09 | | Release date: | 2011-08-31 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Chiral Protein Engineering and its Application in G Health
To be Published
|
|
6NMV
 
 | | Non-Blocking Fab 218 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 218 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 218 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMU
 
 | | Kick-Off Fab 115 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 115 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 115 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMT
 
 | | Non-Blocking Fab 3 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 3 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 3 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
2K1W
 
 | | NMR solution structure of M-crystallin in calcium loaded form(holo). | | Descriptor: | Beta/gama crystallin family protein, CALCIUM ION | | Authors: | Barnwal, R, Jobby, M, Devi, K, Sharma, Y, Chary, K. | | Deposit date: | 2008-03-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of M-crystallin, a primordial betagamma-crystallin from archaea.
J.Mol.Biol., 386, 2009
|
|
2K3Z
 
 | | NMR structure of adenosine bulged RNA duplex with C:G-A triple | | Descriptor: | RNA_(5'-R(*CP*AP*GP*CP*CP*GP*AP*C)-3')_, RNA_(5'-R(*GP*UP*CP*GP*AP*GP*CP*UP*G)-3')_ | | Authors: | Popenda, L, Bielecki, L, Gdaniec, Z, Adamiak, R.W. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple
ARKIVOC, 2009, 2008
|
|
2K40
 
 | |
2K1X
 
 | | NMR solution structure of M-crystallin in calcium free form (apo). | | Descriptor: | Beta/gama crystallin family protein | | Authors: | Barnwal, R, Jobby, M, Devi, K, Sharma, Y, Chary, K. | | Deposit date: | 2008-03-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of M-crystallin, a primordial betagamma-crystallin from archaea.
J.Mol.Biol., 386, 2009
|
|
2JX4
 
 | |
2K3W
 
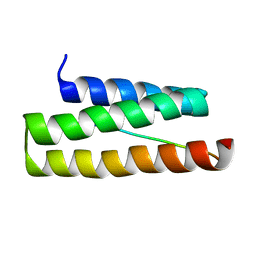 | | NMR structure of VPS4A-MIT-CHMP6 | | Descriptor: | Charged multivesicular body protein 6, Vacuolar protein sorting-associating protein 4A | | Authors: | Kieffer, C, Skalicky, J.J, Morita, E, De Domini, I, Ward, D.M, Kaplan, J, Sundquist, W.I. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding
Dev.Cell, 15, 2008
|
|
2K1D
 
 | |
2JNX
 
 | |
2JOM
 
 | |
2GRI
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2KG5
 
 | | NMR Solution structure of ARAP3-SAM | | Descriptor: | Arf-GAP, Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Leone, M, Pellecchia, M. | | Deposit date: | 2009-03-05 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Sam domain of the lipid phosphatase Ship2 adopts a common model to interact with Arap3-Sam and EphA2-Sam.
Bmc Struct.Biol., 9, 2009
|
|
