4J3E
 
 | | The 1.9A crystal structure of humanized Xenopus Mdm2 with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C.M, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development.
ACS Med Chem Lett, 4, 2013
|
|
4OK5
 
 | |
4IYH
 
 | | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne
To be Published
|
|
7HFW
 
 | |
7HDK
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003730 | | Descriptor: | (2S,3R)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]butane-1,3-diol, CHLORIDE ION, Non-structural protein 3, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
7HEW
 
 | |
7HDP
 
 | |
7HEZ
 
 | |
4HDM
 
 | | Crystal Structure of ArsAB in Complex with p-cresol | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
3G31
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 4 (GF1) | | Descriptor: | (2S)-2-[(3aR,4R,7S,7aS)-1,3-dioxooctahydro-2H-4,7-methanoisoindol-2-yl]propanoic acid, Beta-lactamase CTX-M-9a, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, Y, Shoichet, B.K. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular docking and ligand specificity in fragment-based inhibitor discovery
Nat.Chem.Biol., 5, 2009
|
|
4QMA
 
 | | Crystal Structure of a Putative Cysteine Dioxygnase From Ralstonia eutropha: An Alternative Modeling of 2GM6 from JCSG Target 361076 | | Descriptor: | 1,2-ETHANEDIOL, Cysteine dioxygenase type I, FE (III) ION, ... | | Authors: | Hartman, S.H, Driggers, C.M, Karplus, P.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-06-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of Arg- and Gln-type bacterial cysteine dioxygenase homologs.
Protein Sci., 24, 2015
|
|
5TCA
 
 | | Complement Factor D inhibited with JH3 | | Descriptor: | 1-(2-{(2S)-2-[(6-bromopyridin-2-yl)carbamoyl]-1,3-thiazolidin-3-yl}-2-oxoethyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Complement factor D | | Authors: | Stuckey, J.A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5TB2
 
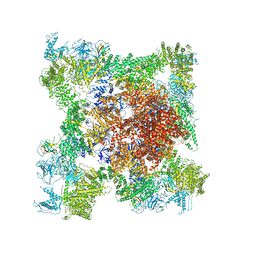 | | Structure of rabbit RyR1 (EGTA-only dataset, class 2) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
7L1S
 
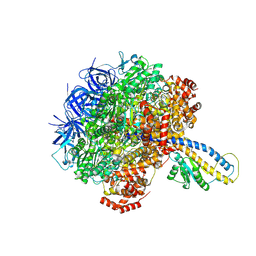 | | PS3 F1-ATPase Pi-bound Dwell | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1Q
 
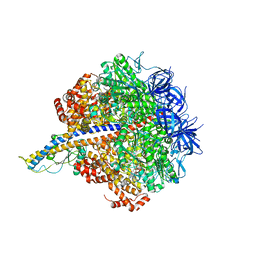 | | PS3 F1-ATPase Binding/TS Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7HFX
 
 | |
7KRI
 
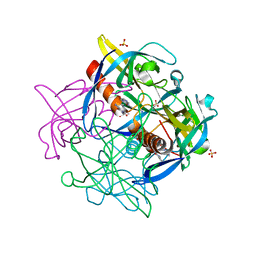 | | FR6-bound SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | 1,3-dimethyl-1H-pyrrolo[3,4-d]pyrimidine-2,4(3H,6H)-dione, MALONATE ION, Non-structural protein 9, ... | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding of a pyrimidine RNA base-mimic to SARS-CoV-2 nonstructural protein 9.
J.Biol.Chem., 297, 2021
|
|
7L1R
 
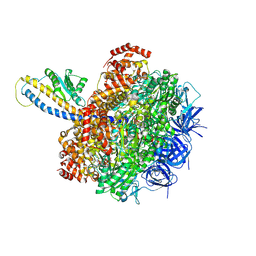 | | PS3 F1-ATPase Hydrolysis Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7DKP
 
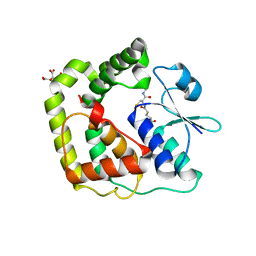 | |
3G97
 
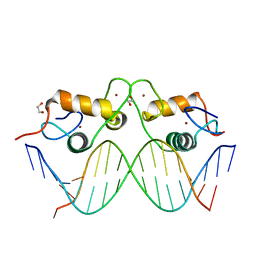 | | GR DNA-binding domain:GilZ 16bp complex-9 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*GP*GP*GP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*CP*CP*CP*AP*AP*TP*GP*TP*TP*CP*T)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
7D8E
 
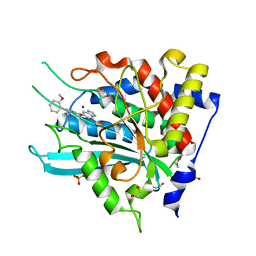 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-09 | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-09
To Be Published
|
|
4B74
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-[(2-ammonioethyl)amino]-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function
Nat.Chem.Biol., 8, 2012
|
|
5TMK
 
 | | Optimization of 3,5-Disubstitued Piperidine: Discovery of Non-Peptide mimetics as an Orally Active Renin Inhibitor | | Descriptor: | 1-(4-methoxybutyl)-N-(2-methylpropyl)-N-[(3S,5R)-5-(morpholine-4-carbonyl)piperidin-3-yl]-5-phenyl-1H-pyrrole-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.C, Lane, W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of 3,5-Disubstitued Piperidine: Discovery of Non-Peptide mimetics as an Orally Active Renin Inhibitor
To be published
|
|
4LWI
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
5TNL
 
 | |
