1EJR
 
 | | CRYSTAL STRUCTURE OF THE D221A VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | NICKEL (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Pearson, M.A, Park, I.S, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-03-04 | | Release date: | 2000-09-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of urease active site variants.
Biochemistry, 39, 2000
|
|
1EJS
 
 | | Crystal Structure of the H219N Variant of Klebsiella Aerogenes Urease | | Descriptor: | NICKEL (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Pearson, M.A, Park, I.S, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-03-04 | | Release date: | 2000-09-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of urease active site variants.
Biochemistry, 39, 2000
|
|
1EJT
 
 | | CRYSTAL STRUCTURE OF THE H219Q VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | NICKEL (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Pearson, M.A, Park, I.S, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-03-04 | | Release date: | 2000-09-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of urease active site variants.
Biochemistry, 39, 2000
|
|
1EJU
 
 | | CRYSTAL STRUCTURE OF THE H320N VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | NICKEL (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Pearson, M.A, Park, I.S, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-03-04 | | Release date: | 2000-09-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of urease active site variants.
Biochemistry, 39, 2000
|
|
1EJV
 
 | | CRYSTAL STRUCTURE OF THE H320Q VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | NICKEL (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Pearson, M.A, Park, I.S, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-03-04 | | Release date: | 2000-09-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and structural characterization of urease active site variants.
Biochemistry, 39, 2000
|
|
1EJW
 
 | |
1EJX
 
 | |
1EJY
 
 | |
1EJZ
 
 | | SOLUTION STRUCTURE OF A HNA-RNA HYBRID | | Descriptor: | DNA (5'-H(*(6HG)P*(6HC)P*(6HG)P*(6HT)P*(6HA)P*(6HG)P*(6HC)P*(6HG))-3'), PHOSPHORIC ACID MONO-(3-HYDROXY-PROPYL) ESTER, RNA (5'-R(*CP*GP*CP*UP*AP*CP*GP*C)-3') | | Authors: | Lescrnier, E, Esnouf, R, Heus, H.A, Hilbers, C.W, Herdewijn, P. | | Deposit date: | 2000-03-06 | | Release date: | 2000-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a HNA-RNA hybrid.
Chem.Biol., 7, 2000
|
|
1EK0
 
 | | GPPNHP-BOUND YPT51 AT 1.48 A RESOLUTION | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Esters, H, Scheidig, A.J. | | Deposit date: | 2000-03-06 | | Release date: | 2000-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | High-resolution crystal structure of S. cerevisiae Ypt51(DeltaC15)-GppNHp, a small GTP-binding protein involved in regulation of endocytosis.
J.Mol.Biol., 298, 2000
|
|
1EK1
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CIU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(4-IODOPHENYL)UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Goodrow, M.H, Dowdy, D.L, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation.
J.Biol.Chem., 275, 2000
|
|
1EK2
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CDU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-DECYLUREA | | Authors: | Argiriadi, M.A, Morisseau, C, Goodrow, M.H, Dowdy, D.L, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation.
J.Biol.Chem., 275, 2000
|
|
1EK3
 
 | | KAPPA-4 IMMUNOGLOBULIN VL, REC | | Descriptor: | CALCIUM ION, CHLORIDE ION, KAPPA-4 IMMUNOGLOBULIN LIGHT CHAIN VL | | Authors: | Pokkuluri, P.R, Huang, D.-B, Raffen, R, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-03-06 | | Release date: | 2001-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Amyloidogenic Kappa-4 Immunoglobulin VL, REC
To be Published
|
|
1EK4
 
 | | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH DODECANOIC ACID TO 1.85 RESOLUTION | | Descriptor: | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I, LAURIC ACID | | Authors: | Olsen, J.G, Kadziola, A, Siggaard-Andersen, M, von Wettstein-Knowles, P, Larsen, S. | | Deposit date: | 2000-03-06 | | Release date: | 2001-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of beta-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery.
Structure, 9, 2001
|
|
1EK5
 
 | | STRUCTURE OF HUMAN UDP-GALACTOSE 4-EPIMERASE IN COMPLEX WITH NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE 4-EPIMERASE | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase.
Biochemistry, 39, 2000
|
|
1EK6
 
 | | STRUCTURE OF HUMAN UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH NADH AND UDP-GLUCOSE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, TETRAMETHYLAMMONIUM ION, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase.
Biochemistry, 39, 2000
|
|
1EK8
 
 | |
1EK9
 
 | | 2.1A X-RAY STRUCTURE OF TOLC: AN INTEGRAL OUTER MEMBRANE PROTEIN AND EFFLUX PUMP COMPONENT FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PROTEIN TOLC | | Authors: | Koronakis, V, Sharff, A.J, Koronakis, E, Luisi, B, Hughes, C. | | Deposit date: | 2000-03-07 | | Release date: | 2000-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export.
Nature, 405, 2000
|
|
1EKA
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*GP*CP*UP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
1EKB
 
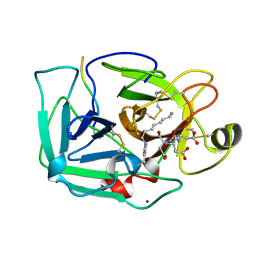 | | THE SERINE PROTEASE DOMAIN OF ENTEROPEPTIDASE BOUND TO INHIBITOR VAL-ASP-ASP-ASP-ASP-LYS-CHLOROMETHANE | | Descriptor: | ENTEROPEPTIDASE, VAL-ASP-ASP-ASP-ASP-LYK PEPTIDE, ZINC ION | | Authors: | Fuetterer, K, Lu, D, Sadler, J.E, Waksman, G. | | Deposit date: | 1999-05-02 | | Release date: | 1999-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enteropeptidase light chain complexed with an analog of the trypsinogen activation peptide.
J.Mol.Biol., 292, 1999
|
|
1EKD
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
1EKE
 
 | | CRYSTAL STRUCTURE OF CLASS II RIBONUCLEASE H (RNASE HII) WITH MES LIGAND | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, RIBONUCLEASE HII | | Authors: | Lai, L.H, Yokota, H, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-03-07 | | Release date: | 2000-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of archaeal RNase HII: a homologue of human major RNase H
Structure, 8, 2000
|
|
1EKF
 
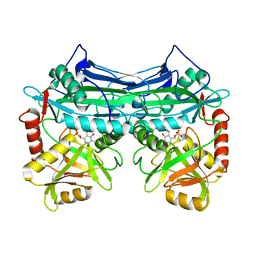 | | CRYSTALLOGRAPHIC STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 1.95 ANGSTROMS (ORTHORHOMBIC FORM) | | Descriptor: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-08 | | Release date: | 2001-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EKG
 
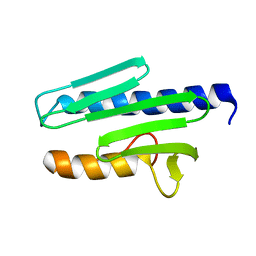 | | MATURE HUMAN FRATAXIN | | Descriptor: | FRATAXIN | | Authors: | Dhe-Paganon, S, Shigeta, R, Chi, Y.I, Ristow, M, Shoelson, S.E. | | Deposit date: | 2000-03-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human frataxin.
J.Biol.Chem., 275, 2000
|
|
1EKH
 
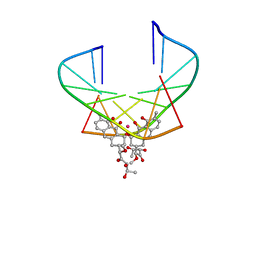 | | NMR STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
