4W9G
 
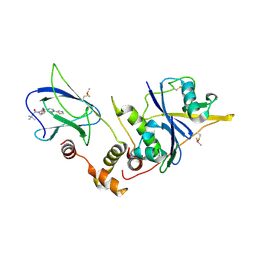 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(3-methyl-4-(thiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[3-methyl-4-(1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4W9D
 
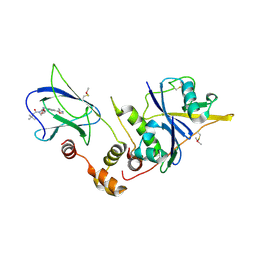 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methyloxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
1EER
 
 | |
7EGP
 
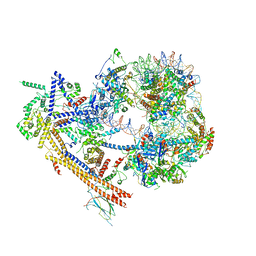 | | The structure of SWI/SNF-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP9, Actin-related protein 7, ... | | Authors: | Chen, Z.C, Chen, K.J, He, Z.Y, Ye, Y.P. | | Deposit date: | 2021-03-24 | | Release date: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the SWI/SNF complex bound to the nucleosome and insights into the functional modularity.
Cell Discov, 7, 2021
|
|
1EYK
 
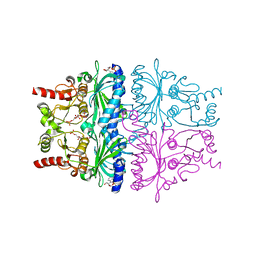 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH AMP, ZINC, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
1EYJ
 
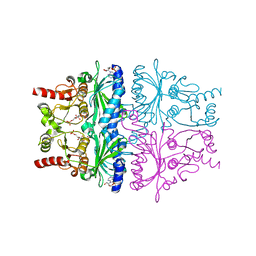 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH AMP, MAGNESIUM, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
1EYI
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH MAGNESIUM, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (R-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE-1,6-BISPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-06 | | Release date: | 2000-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
2HYU
 
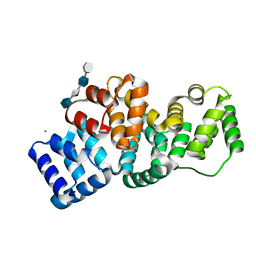 | | Human Annexin A2 with heparin tetrasaccharide bound | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Annexin A2, CALCIUM ION | | Authors: | Shao, C, Head, J.F, Seaton, B.A. | | Deposit date: | 2006-08-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic Analysis of Calcium-dependent Heparin Binding to Annexin A2.
J.Biol.Chem., 281, 2006
|
|
2HYW
 
 | | Human Annexin A2 with Calcium bound | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Shao, C, Head, J.F, Seaton, B.A. | | Deposit date: | 2006-08-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Analysis of Calcium-dependent Heparin Binding to Annexin A2.
J.Biol.Chem., 281, 2006
|
|
7TAN
 
 | | Structure of VRK1 C-terminal tail bound to nucleosome core particle | | Descriptor: | Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, Histone H3.2, ... | | Authors: | Spangler, C.J, Budziszewski, G.R, McGinty, R.K. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multivalent DNA and nucleosome acidic patch interactions specify VRK1 mitotic localization and activity.
Nucleic Acids Res., 50, 2022
|
|
6RNY
 
 | | PFV intasome - nucleosome strand transfer complex | | Descriptor: | DNA (108-MER), DNA (128-MER), DNA (33-MER), ... | | Authors: | Pye, V.E, Renault, L, Maskell, D.P, Cherepanov, P, Costa, A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
7XJG
 
 | | Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom | | Descriptor: | DNA (105-MER), MAGNESIUM ION, RNA (14-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2022-04-17 | | Release date: | 2022-09-14 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
6MSJ
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSG
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSE
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
2EXU
 
 | | Crystal Structure of Saccharomyces cerevisiae transcription elongation factors Spt4-Spt5NGN domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOL, Transcription initiation protein SPT4/SPT5, ... | | Authors: | Xu, F, Guo, M, Fang, P, Teng, M, Niu, L. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae transcription elongation factors Spt4-Spt5NGN domain
To be published
|
|
5YDE
 
 | | Crystal structure of a disease-related gene, hCDC73(1-111) | | Descriptor: | Parafibromin | | Authors: | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.023 Å) | | Cite: | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
9MGN
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 41 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclobutylamino)-N-[(2S)-2-hydroxy-3-{6-[(1H-pyrazol-4-yl)methoxy]-3,4-dihydroisoquinolin-2(1H)-yl}propyl]pyridine-4-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9MGM
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 24 | | Descriptor: | 2-(cyclobutylamino)-N-[(2S)-2-hydroxy-3-{6-[(1-methyl-1H-pyrazol-5-yl)methoxy]-3,4-dihydroisoquinolin-2(1H)-yl}propyl]pyridine-4-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9MGR
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 51 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 6-[(1-acetylazetidin-3-yl)amino]-N-[(2R)-2-hydroxy-2-{(3S)-7-[(4-methyl-1,3-oxazol-5-yl)methoxy]-1,2,3,4-tetrahydroisoquinolin-3-yl}ethyl]-2-(4-methylpiperidin-1-yl)pyrimidine-4-carboxamide, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9MGQ
 
 | | Crystal structure of PRMT5:MEP50 in complex with sinefungin and compound 47 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclobutylamino)-N-{(2R)-2-hydroxy-2-[(3S)-1,2,3,4-tetrahydroisoquinolin-3-yl]ethyl}pyridine-4-carboxamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9MGP
 
 | |
9MGL
 
 | |
5YJ8
 
 | |
5YBC
 
 | | X-ray structure of native ETS-domain domain of Ergp55 | | Descriptor: | GLYCEROL, Transcriptional regulator ERG | | Authors: | Saxena, A.K, Gangwar, S.P. | | Deposit date: | 2017-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparative structure analysis of the ETSi domain of ERG3 and its complex with the E74 promoter DNA sequence
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
