8J8A
 
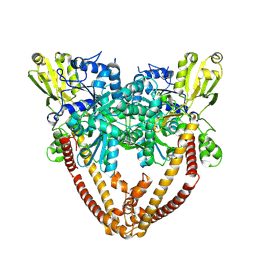 | |
6TUR
 
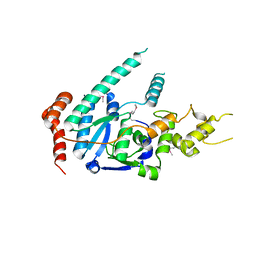 | | human XPG, Apo1 form | | Descriptor: | DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
7XFN
 
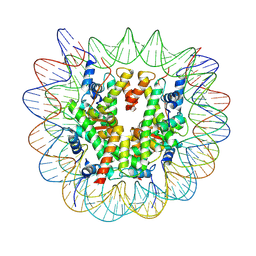 | | Structure of nucleosome-DI complex (-55I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFC
 
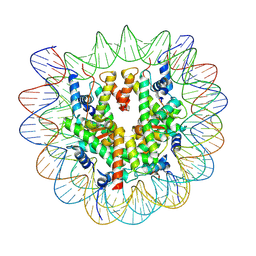 | | Structure of nucleosome-DI complex (-30I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFI
 
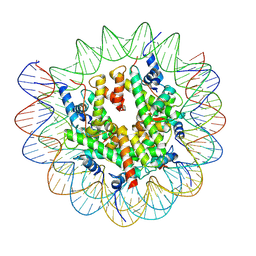 | | Structure of nucleosome-DI complex (-50I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFL
 
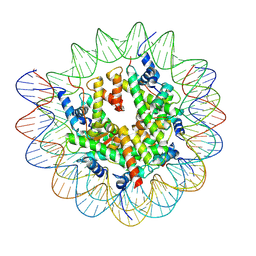 | | Structure of nucleosome-AAG complex (A-53I, free state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XNP
 
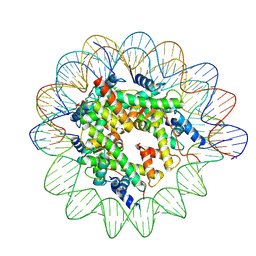 | | Structure of nucleosome-AAG complex (A-55I, post-catalytic state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
6TUS
 
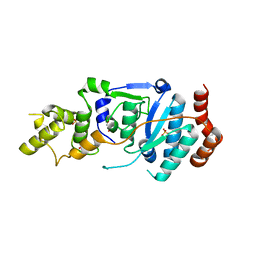 | | human XPG, Apo2 form | | Descriptor: | DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells, GLYCEROL, SULFATE ION | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
7M99
 
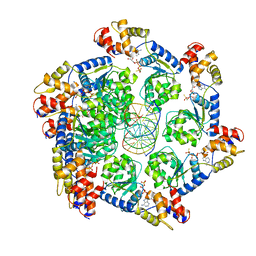 | | ATPgS bound TnsC filament from ShCAST system | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
7OOP
 
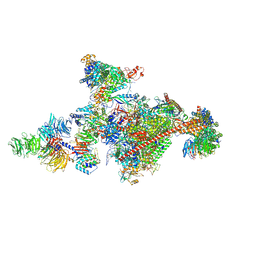 | | Pol II-CSB-CSA-DDB1-UVSSA-PAF-SPT6 (Structure 3) | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
9C8V
 
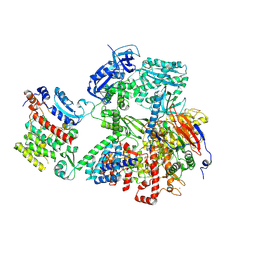 | | Human DNA polymerase alpha/primase - CHAPSO (4 mM) | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Abe, K.M, Li, G, He, Q, Grant, T, Lim, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Human DNA polymerase alpha/primase - CHAPSO (4 mM)
To Be Published
|
|
9CEW
 
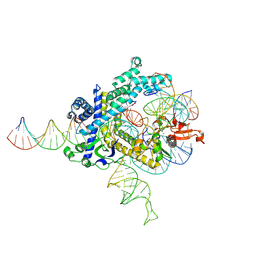 | | Spizellomyces punctatus Fanzor (SpuFz) State 3 | | Descriptor: | DNA (29-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9BAQ
 
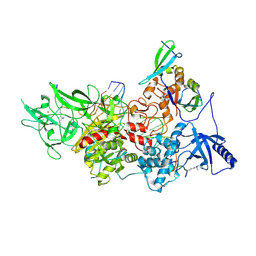 | | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*CP*T)-R(P*(PYO))-D(P*CP*TP*CP*CP*TP*CP*CP*TP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*TP*AP*GP*T)-3'), DNA (cytosine-5-)-methyltransferase, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex
To Be Published
|
|
9CEX
 
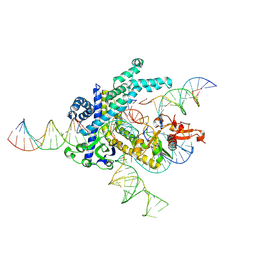 | | Spizellomyces punctatus Fanzor (SpuFz) State 4 | | Descriptor: | DNA (29-MER), DNA (5'-D(*(MG)*(MG)P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
8DRH
 
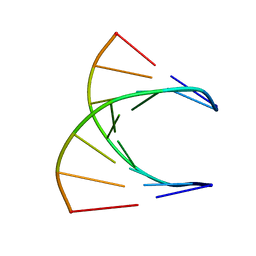 | | HIGH RESOLUTION NMR STRUCTURE OF THE D(GCGTCAGG)R(CCUGACGC) HYBRID, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*CP*AP*GP*G)-3'), RNA (5'-R(*CP*CP*UP*GP*AP*CP*GP*C)-3') | | Authors: | Bachelin, M, Hessler, G, Kurz, G, Hacia, J.G, Dervan, P.B, Kessler, H. | | Deposit date: | 1997-10-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stereoregular Phosphorothioate DNA/RNA Duplex
Nat.Struct.Biol., 5, 1998
|
|
9CF2
 
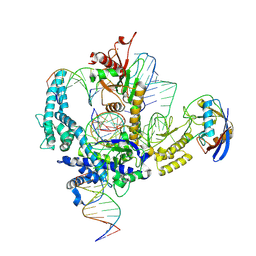 | | Parasitella parasitica Fanzor (PpFz) State 3 | | Descriptor: | DNA non-target strand, DNA substrate model, DNA target strand, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
2PJR
 
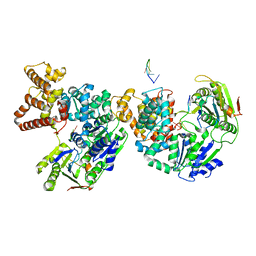 | | HELICASE PRODUCT COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*T)-3'), ... | | Authors: | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
9GSQ
 
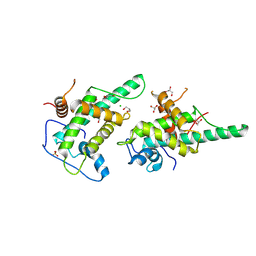 | | DNA binding domain of J-DNA Binding Protein 3 (JBP3) | | Descriptor: | CHLORIDE ION, DNA binding domain of J-DNA binding protein 3, GLYCEROL, ... | | Authors: | de Vries, I, Adamopoulos, A, Joosten, R.P, Perrakis, A. | | Deposit date: | 2024-09-16 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Base-J binding proteins JBP1 and JBP3 have conserved structures of their J-DNA binding domain but drastically different affinities
To Be Published
|
|
6EL8
 
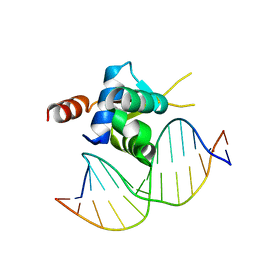 | | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*GP*CP*GP*TP*CP*TP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*AP*GP*AP*CP*GP*CP*CP*AP*CP*C)-3'), Forkhead box protein N1 | | Authors: | Newman, J.A, Aitkenhead, H.A, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA
To be published
|
|
9CF3
 
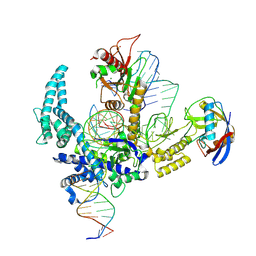 | | Parasitella parasitica Fanzor (PpFz) State 4 | | Descriptor: | DNA non-target strand, DNA target strand, MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEV
 
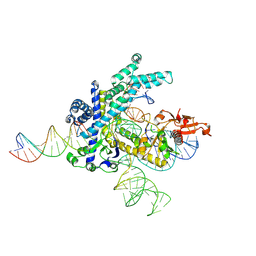 | | Spizellomyces punctatus Fanzor (SpuFz) State 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF1
 
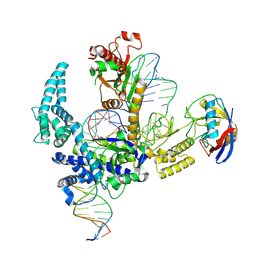 | | Parasitella parasitica Fanzor (PpFz) State 2 | | Descriptor: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CES
 
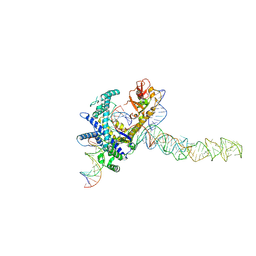 | | Guillardia theta Fanzor (GtFz) State 2 | | Descriptor: | DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*GP*GP*GP*CP*CP*TP*TP*TP*AP*AP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF0
 
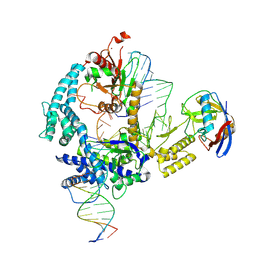 | | Parasitella parasitica Fanzor (PpFz) State 1 | | Descriptor: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CET
 
 | | Guillardia theta Fanzor (GtFz) State 3 | | Descriptor: | DNA (28-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
