7ZO3
 
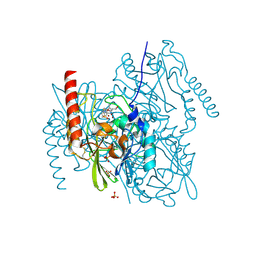 | | L1 metallo-beta-lactamase in complex with hydrolysed tebipenem | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[1-(4,5-dihydro-1,3-thiazol-2-yl)azetidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
9D7R
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with Fva1 antimicrobial peptide, mRNA, A-site release factor 1, and deacylated P-site and E-site tRNAphe at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Huang, W, Baliga, C, Atkinson, G.C, Vazquez-Laslop, N, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-17 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activity, structure, and diversity of Type II proline-rich antimicrobial peptides from insects.
Embo Rep., 25, 2024
|
|
7ZO4
 
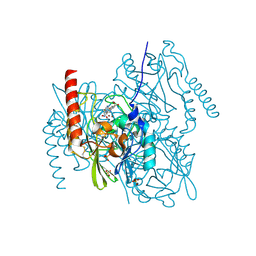 | | L1 metallo-beta-lactamase in complex with hydrolysed panipenem | | Descriptor: | (2R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S)-1-ethanimidoylpyrrolidin-3-yl]sulfanyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SODIUM ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
5OO8
 
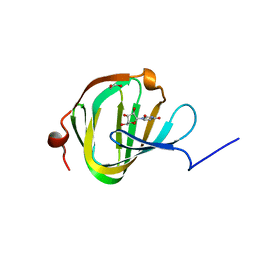 | | Streptomyces PAC13 (H42Q) with uridine | | Descriptor: | 1,2-ETHANEDIOL, Putative cupin_2 domain-containing isomerase, URIDINE | | Authors: | Chung, C, Michailidou, F. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6KU0
 
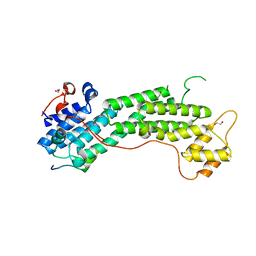 | | Crystal structure of MyoVa-GTD in complex with MICAL1-GTBM | | Descriptor: | 1,2-ETHANEDIOL, Peptide from [F-actin]-monooxygenase MICAL1, Unconventional myosin-Va | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | F-actin disassembly factor MICAL1 binding to Myosin Va mediates cargo unloading during cytokinesis.
Sci Adv, 6, 2020
|
|
5AKG
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 5-(2-thienyl)-1,3,4-thiadiazol-2-amine, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
6AT5
 
 | |
153L
 
 | |
4A60
 
 | | Crystal structure of human testis-specific fatty acid binding protein 9 (FABP9) | | Descriptor: | 1,2-ETHANEDIOL, FATTY ACID-BINDING PROTEIN 9 TESTIS LIPID-BINDING PROTEIN, TLBP, ... | | Authors: | Muniz, J.R.C, Kiyani, W, Shrestha, L, Froese, D.S, Krojer, T, Vollmar, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, von Delft, F, Yue, W.W. | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Crystal Structure of the Human Fabp9A
To be Published
|
|
6KX4
 
 | |
5ALG
 
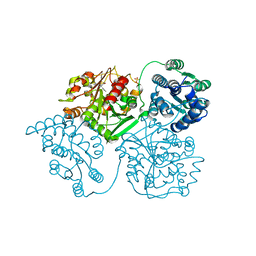 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 4-[(3-chlorophenyl)methyl]-N-[[(3S)-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]-3-oxidanylidene-1,4-benzothiazine-6-carboxamide, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
7RD2
 
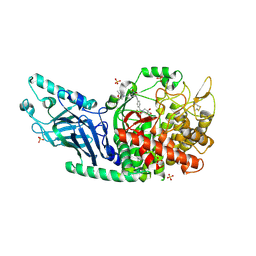 | | Co-crystal structure of Chaetomium glucosidase with compound 2 | | Descriptor: | (2R,3R,4R,5S)-1-{[4-({4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-2-nitroanilino}methyl)phenyl]methyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-09 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6QMF
 
 | |
5HTI
 
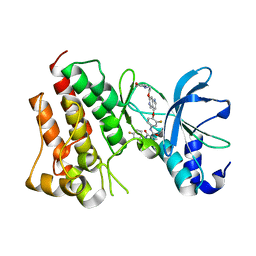 | | Crystal structure of c-Met kinase domain in complex with LXM108 | | Descriptor: | Hepatocyte growth factor receptor, N-[3-fluoro-4-({7-[2-(morpholin-4-yl)ethoxy]-1,6-naphthyridin-4-yl}oxy)phenyl]-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of c-Met kinase domain in complex with LXM108
to be published
|
|
5DGH
 
 | |
6HTY
 
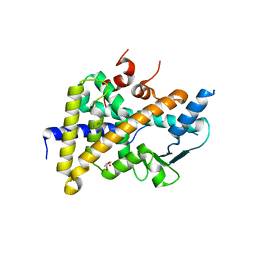 | | PXR in complex with P2X4 inhibitor compound 25 | | Descriptor: | (2~{R})-~{N}-[4-(3-chloranylphenoxy)-3-sulfamoyl-phenyl]-2-phenyl-propanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Puetter, V, Werner, S, Mesch, S, Laux-Biehlmann, A, Braeuer, N, Dahloef, H, Klint, J, ter Laak, A, Pook, E, Neagoe, I, Nubbemeyer, R, Schulz, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and Characterization of the Potent and Selective P2X4 InhibitorN-[4-(3-Chlorophenoxy)-3-sulfamoylphenyl]-2-phenylacetamide (BAY-1797) and Structure-Guided Amelioration of Its CYP3A4 Induction Profile.
J.Med.Chem., 62, 2019
|
|
7JM3
 
 | | Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer | | Descriptor: | Matrix protein 1 | | Authors: | Su, Z, Pintilie, G, Selzer, L, Chiu, W, Kirkegaard, K. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer.
Plos Biol., 18, 2020
|
|
3IHB
 
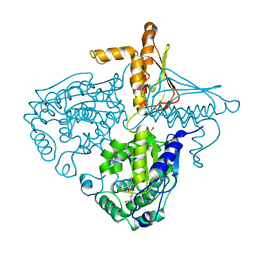 | | Crystal Structure Analysis of Mglu in its tris and glutamate form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-29 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product l-glutamate and its activator Tris
Febs J., 277, 2010
|
|
7JMH
 
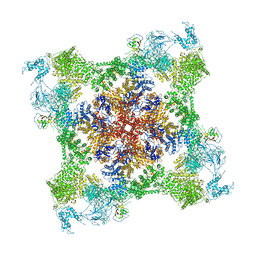 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 35 - State 4 (S4) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
6AYR
 
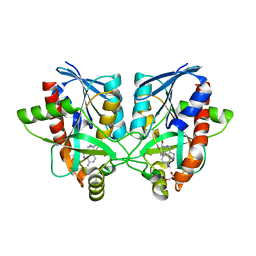 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
7CDY
 
 | |
5KPQ
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-propyl-piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
6EUH
 
 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, Beta-glucanase, CALCIUM ION | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
5HLB
 
 | | E. coli PBP1b in complex with acyl-aztreonam and moenomycin | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Escherichia coli Penicillin-Binding Protein 1B: Structural Insights into Inhibition.
J. Biol. Chem., 2016
|
|
8EAZ
 
 | | HOIL-1/E2-Ub/Ub transthiolation complex | | Descriptor: | RanBP-type and C3HC4-type zinc finger-containing protein 1, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Wang, X.S, Cotton, T.R, Lechtenberg, B.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family.
Nat Commun, 14, 2023
|
|
