1RN7
 
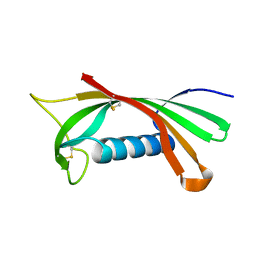 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
2PK3
 
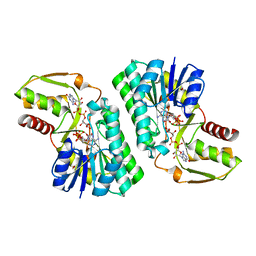 | | Crystal Structure of a GDP-4-keto-6-deoxy-D-mannose reductase | | Descriptor: | GDP-6-deoxy-D-lyxo-4-hexulose reductase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5R)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Webb, N.A, Garavito, R.M. | | Deposit date: | 2007-04-17 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The structural basis for catalytic function of GMD and RMD, two closely related enzymes from the GDP-D-rhamnose biosynthesis pathway.
Febs J., 276, 2009
|
|
1UXB
 
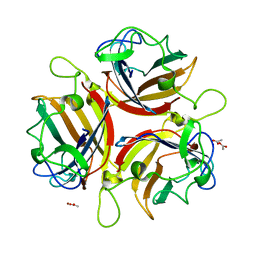 | | ADENOVIRUS AD19p FIBRE HEAD in complex with sialyl-lactose | | Descriptor: | ACETATE ION, FIBER PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
1ROA
 
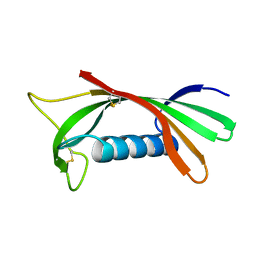 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1DIE
 
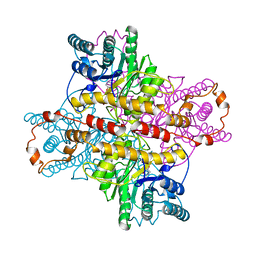 | | OBSERVATIONS OF REACTION INTERMEDIATES AND THE MECHANISM OF ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE | | Descriptor: | 1-DEOXYNOJIRIMYCIN, D-XYLOSE ISOMERASE, MAGNESIUM ION | | Authors: | Collyer, C.A, Viehmann, H, Goldberg, J.D, Blow, D.M. | | Deposit date: | 1992-06-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observations of reaction intermediates and the mechanism of aldose-ketose interconversion by D-xylose isomerase.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
2EGH
 
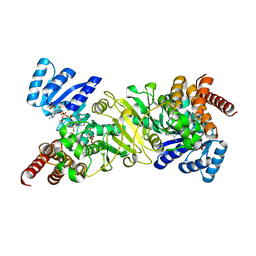 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with a magnesium ion, NADPH and fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | Authors: | Yajima, S, Hara, K, Iino, D, Sasaki, Y, Kuzuyama, T, Seto, H. | | Deposit date: | 2007-03-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in a quaternary complex with a magnesium ion, NADPH and the antimalarial drug fosmidomycin
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2EPO
 
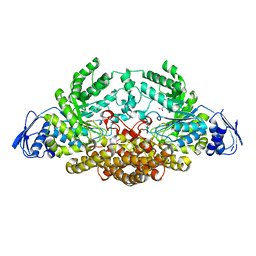 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | ACETIC ACID, N-acetyl-beta-D-glucosaminidase | | Authors: | Langley, D.B. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
1TOQ
 
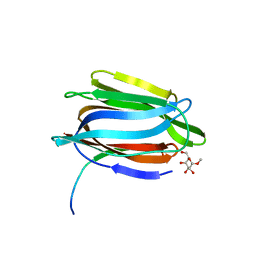 | | CRYSTAL STRUCTURE OF A GALACTOSE SPECIFIC LECTIN FROM ARTOCARPUS HIRSUTA IN COMPLEX WITH METHYL-a-D-GALACTOSE | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA CHAIN, methyl alpha-D-galactopyranoside | | Authors: | Rao, K.N, Suresh, C.G, Katre, U.V, Gaikwad, S.M, Khan, M.I. | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two orthorhombic crystal structures of a galactose-specific lectin from Artocarpus hirsuta in complex with methyl-alpha-D-galactose.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2A2G
 
 | | THE CRYSTAL STRUCTURES OF A2U-GLOBULIN AND ITS COMPLEX WITH A HYALINE DROPLET INDUCER. | | Descriptor: | D-LIMONENE 1,2-EPOXIDE, PROTEIN (ALPHA-2U-GLOBULIN) | | Authors: | Chaudhuri, B.N, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 1998-11-19 | | Release date: | 1999-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structures of alpha 2u-globulin and its complex with a hyaline droplet inducer.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1UNM
 
 | | Crystal structure of 7-Aminoactinomycin D with non-complementary DNA | | Descriptor: | 5'-D(*TP*TP*AP*GP*BRU*TP)-3', 7-AMINOACTINOMYCIN D | | Authors: | Alexopoulos, E.C, Klement, R, Jares-Erijman, E.A, Uson, I, Jovin, T.M, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and Solution Structures of 7-Amino-Actinomycin D Complexes with D(Ttagbrut), D(Ttagtt) and D(Tttagttt)
Acta Crystallogr.,Sect.D, 61, 2005
|
|
326D
 
 | | CRYSTAL STRUCTURES OF D(CM5CGGGCCM5CGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*(5CM)P*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
1HFD
 
 | | HUMAN COMPLEMENT FACTOR D IN A P21 CRYSTAL FORM | | Descriptor: | COMPLEMENT FACTOR D | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
322D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCM5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
321D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCCGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
318D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCC(BR)5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(CBR)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
320D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCCGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
319D
 
 | | CRYSTAL STRUCTURES OF D(CCGGG(BR)5CCCGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*(CBR)P*CP*CP*GP*G)-3'), THERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
2EPL
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | GLYCEROL, N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2EPM
 
 | | N-acetyl-B-D-glucoasminidase (GCNA) from Stretococcus gordonii | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-acetyl-beta-D-glucosaminidase, ... | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2EPK
 
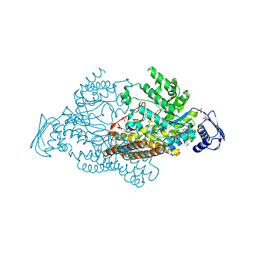 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2F9R
 
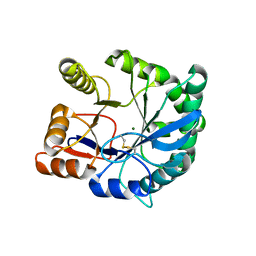 | | Crystal structure of the inactive state of the Smase I, a sphingomyelinase D from Loxosceles laeta venom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Sphingomyelinase D 1 | | Authors: | Murakami, M.T, Gabdoulkhakov, A, Fernandes-Pedrosa, M.F, Betzel, C, Tambourgi, D.V, Arni, R.K. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases D.
J.Biol.Chem., 280, 2005
|
|
3P93
 
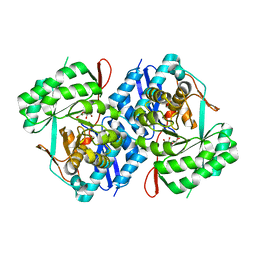 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter Salexigens complexed with MG,D-Mannonate and 2-keto-3-deoxy-D-Gluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, D-MANNONIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-15 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-MANNONATE DEHYDRATASE FROM CHROMOHALOBACTER SALEXIGENS complexed with MG,D-Mannonate and 2-keto-3-deoxy-D-Gluconate
To be Published
|
|
4E52
 
 | |
325D
 
 | | CRYSTAL STRUCTURES OF D(CM5CGGGCCM5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*(5CM)P*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
