9HJH
 
 | | Structure of compound 1 bound to SARS-CoV-2 main protease | | Descriptor: | (2~{R})-4-[(4-bromanyl-2-ethyl-phenyl)methyl]-1-(5-chloranylpyridin-3-yl)carbonyl-~{N}-ethyl-1,4-diazepane-2-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2024-11-29 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Accelerating the Hit-To-Lead Optimization of a SARS-CoV-2 Mpro Inhibitor Series by Combining High-Throughput Medicinal Chemistry and Computational Simulations.
J.Med.Chem., 68, 2025
|
|
4ZX8
 
 | |
6FTA
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-3098 | | Descriptor: | (4~{a}~{S},8~{a}~{S})-4-[4-methoxy-3-[(2-methoxyphenyl)methoxy]phenyl]-2-[1-(3-nitroimidazo[1,2-b]pyridazin-6-yl)piperidin-4-yl]-4~{a},5,6,7,8,8~{a}-hexahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-3098
To be published
|
|
5CF6
 
 | | CRYSTAL STRUCTURE OF JANUS KINASE 2 IN COMPLEX WITH N,N-DICYCLOPROPYL-10-[(2S)-2,3-DIHYDROXYPROPYL]-3-METHYL-7-(METHYLAMINO)-3,5,8,10-TETRAAZATRICYCLO [7.3.0.02,6]DODECA-1(9),2(6),4,7,11-PENTAENE-11-CARBOXAMIDE | | Descriptor: | N,N-dicyclopropyl-6-[(2S)-2,3-dihydroxypropyl]-1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
1Y44
 
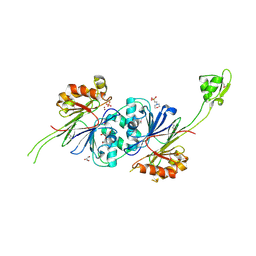 | | Crystal structure of RNase Z | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | de la Sierra-Gallay, I.L, Pellegrini, O, Condon, C. | | Deposit date: | 2004-11-30 | | Release date: | 2005-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z.
Nature, 433, 2005
|
|
5VNB
 
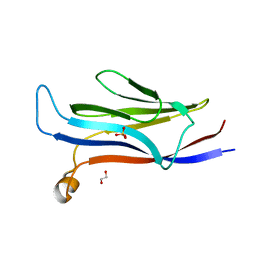 | | YEATS in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, H3K23acK27ac peptide, SULFATE ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2017-04-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode.
ACS Chem. Biol., 13, 2018
|
|
4TW6
 
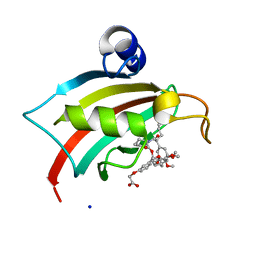 | | The Fk1 domain of FKBP51 in complex with iFit1 | | Descriptor: | (3-{(1R)-3-(3,4-dimethoxyphenyl)-1-[({(2S)-1-[(2S)-2-(3,4,5-trimethoxyphenyl)pent-4-enoyl]piperidin-2-yl}carbonyl)oxy]propyl}phenoxy)acetic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
6RGD
 
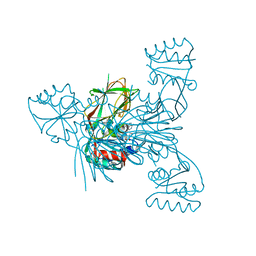 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[4-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]but-3-ynylamino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
7D61
 
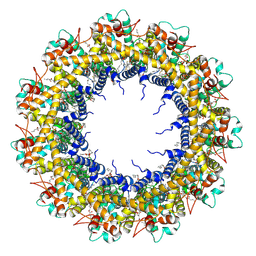 | | Cryo-EM Structure of human CALHM5 in the presence of EDTA | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
8HFY
 
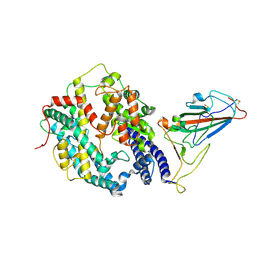 | | SARS-CoV-2 Omicron BA.1 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
7XWZ
 
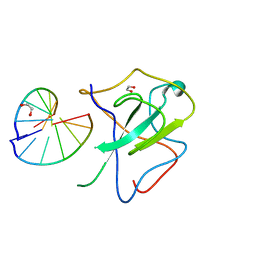 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
4U15
 
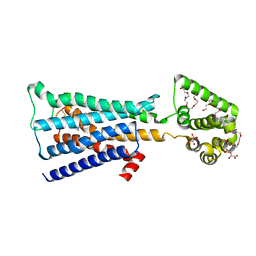 | | M3-mT4L receptor bound to tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, D(-)-TARTARIC ACID, ... | | Authors: | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | Deposit date: | 2014-07-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
5KNY
 
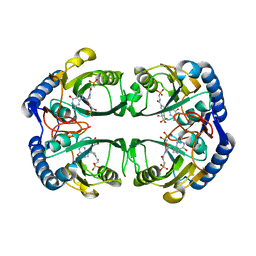 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with (3-((3R,4R)-3-(Guanin-9-yl)-4-((S)-2-hydroxy-2-phosphonoethoxy)pyrrolidin-1-yl)-3-oxopropyl)phosphonic acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
5AAL
 
 | | Complex of the FimH lectin with a C-linked para-biphenyl ethylene alpha-D-mannoside in soaked trigonal crystals at 2.45 A resolution | | Descriptor: | (2R,3S,4R,5S,6R)-2-(hydroxymethyl)-6-[(Z)-3-(4-phenylphenyl)prop-2-enyl]oxane-3,4,5-triol, FIMH, NICKEL (II) ION, ... | | Authors: | De Ruyck, J, Bouckaert, J. | | Deposit date: | 2015-07-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of C-Mannosylated Anti-Adhesives Bound to the Type 1 Fimbrial Fimh Adhesin
Iucrj, 3, 2016
|
|
4TWC
 
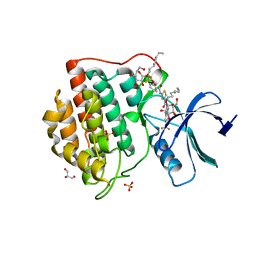 | | 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4- carboxamide derivatives as potent inhibitors of CK1d/e | | Descriptor: | 2-{[2-(trifluoromethoxy)benzoyl]amino}-N-[6-(trifluoromethyl)-1H-benzimidazol-2-yl]-1,3-thiazole-4-carboxamide, Casein kinase I isoform delta, DIMETHYL SULFOXIDE, ... | | Authors: | Bischof, J, Leban, L, Zaja, M, Grothey, A, Radunsky, B, Othersen, O, Strobl, S, Vitt, D, Knippschild, U. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1d/e.
Amino Acids, 43, 2012
|
|
5KPO
 
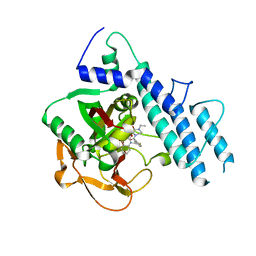 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[3-(4-ethyl-3-oxidanylidene-piperazin-1-yl)carbonyl-4-fluoranyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5HCZ
 
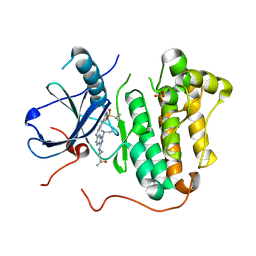 | | EGFR kinase domain mutant "TMLR" with 3-azetidinyl azaindazole compound 21 | | Descriptor: | 2-[1-[1-[(2~{S})-butan-2-yl]-6-[[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]amino]pyrazolo[4,3-c]pyridin-3-yl]azetidin-3-yl]propan-2-ol, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of a Noncovalent, Mutant-Selective Epidermal Growth Factor Receptor Inhibitor.
J.Med.Chem., 59, 2016
|
|
6RAV
 
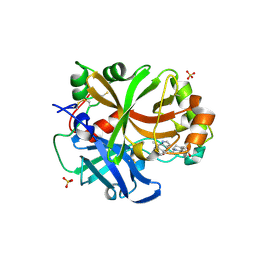 | | Complement factor B protease domain in complex with the reversible inhibitor 4-((2S,4S)-4-ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 4-[(2~{S},4~{S})-4-ethoxy-1-[(5-methoxy-7-methyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
9MRE
 
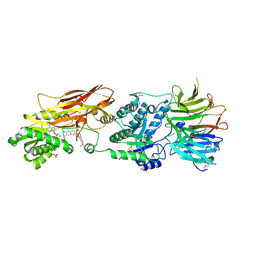 | |
9NAI
 
 | | LuIII VLP - Glycan [s(Lex)2] | | Descriptor: | Capsid protein VP1, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Busuttil, K.B, Bennett, A.B. | | Deposit date: | 2025-02-12 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Mapping the Sialic Acid Binding Sites of LuIII and H-1 Parvovirus
To Be Published
|
|
6EKJ
 
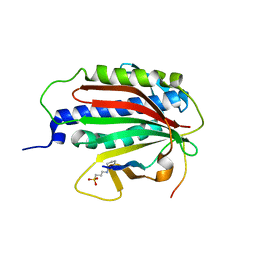 | |
5VKQ
 
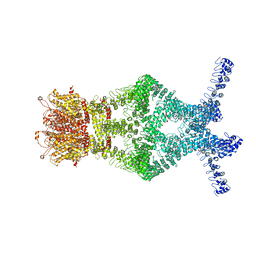 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | Authors: | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | Deposit date: | 2017-04-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
4U71
 
 | |
9HCE
 
 | | Mouse mitoribosome large subunit assembly intermediate (without uL16m) bound to MRM3 dimer and the MALSU-L0R8F8-mt-ACP complex, State A3 (SAMC knock out) | | Descriptor: | 16S rRNA (1584-MER), Acyl carrier protein, mitochondrial, ... | | Authors: | Singh, V, Rorbach, J, Freyer, C, Amunts, A, Wredenberg, A. | | Deposit date: | 2024-11-08 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (7.31 Å) | | Cite: | The mitochondrial methylation potential gates mitoribosome assembly.
Nat Commun, 16, 2025
|
|
6RY0
 
 | | Crystal structure of Dfg5 from Chaetomium thermophilum | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Mannan endo-1,6-alpha-mannosidase, ... | | Authors: | Essen, L.-O, Vogt, M.S. | | Deposit date: | 2019-06-10 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural base for the transfer of GPI-anchored glycoproteins into fungal cell walls.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
